Clinical utility of postablation liver tumor biopsy and possibility of gene mutation analysis
Abstract
Aim
Radiofrequency ablation (RFA) is regarded as a first-line treatment for hepatocellular carcinoma (HCC) at an early stage. When treated with RFA, tumor biopsy may not be performed due to the risk of neoplastic seeding. We previously revealed that the risk of neoplastic seeding is significantly reduced by performing biopsies after RFA. In this study, we investigated the possibility of pathological evaluation and gene mutation analysis of post-RFA tumor specimens.
Methods
Radiofrequency ablation was undertaken on diethylnitrosamine-induced mouse liver tumor, and tumor samples with or without RFA were subjected to whole exome sequencing. Post-RFA human liver tumor specimens were used for detection of TERT promoter mutations and pathological assessment.
Results
The average somatic mutation rate, sites of mutation, and small indels and base transition patterns were comparable between the nontreated and post-RFA tumors. We identified 684 sites of nonsynonymous somatic substitutions in the nontreated tumor and 704 sites of nonsynonymous somatic substitutions in the post-RFA tumor, with approximately 85% in common. In the human post-RFA samples, the TERT promoter mutations were successfully detected in 40% of the cases. Pathological evaluation was possible with post-RFA specimens, and in one case, the diagnosis of adenocarcinoma was made.
Conclusion
Our findings suggest that post-RFA liver tumor biopsy is a useful and safe method for obtaining tumor samples that can be used for gene mutation analysis and for pathological assessment.
Abbreviations
-
- CT
-
- computed tomography
-
- H&E
-
- hematoxylin and eosin
-
- HCC
-
- hepatocellular carcinoma
-
- ICC
-
- intrahepatic cholangiocarcinoma
-
- RFA
-
- radiofrequency ablation
-
- SNV
-
- single nucleotide variant
-
- TERT
-
- telomerase reverse transcriptase
INTRODUCTION
In recent years, systemic therapy for advanced HCC has undergone remarkable development with the introduction of various molecular-targeted agents and immune checkpoint inhibitors.1, 2 These advancements have a profound impact on the management of patients with advanced HCC. In line with this, there is an urgent need to predict the therapeutic efficacy and side-effects to select appropriate agents for the cost-effectiveness of treatment and preservation of liver function.3-5 Against this background, precision medicine based on genetic tumor mutations is expected to be the future of HCC treatment.
The collection of tumor specimens is an essential step in the quest for an effective cancer genomic medicine. However, neoplastic seeding can be a serious problem in needle biopsies of HCC, leading to poor prognosis.6, 7 We have previously reported that tumor biopsies prior to RFA can cause subsequent neoplastic seeding.8 Therefore, in cases of HCC that can be cured by RFA, pathological diagnosis may not be performed. However, if the ablated tumor is not actually HCC but, for example, ICC, the patient may suffer disadvantages, such as lack of appropriate postoperative management. Furthermore, in cases where advanced HCC has recurred after treatment with RFA, tumor samples might not be available because of a high risk of bleeding due to compromised liver function or technical difficulties with puncture due to the localization of the tumor. In such cases, the opportunity to benefit from genomic medicine is lost.
We have previously reported that conducting tumor biopsies after RFA drastically reduces the risk of the neoplastic seeding.9 Therefore, post-RFA tumor biopsy can be a useful and safe method for collecting tumor samples. If pathological assessment could be performed on post-RFA specimens, appropriate postoperative management would be possible, especially when liver tumors with atypical radiological imaging are ablated. In addition, if gene mutation analysis could be performed on the collected post-RFA specimens, the feasibility of genomic medicine would increase in future adjuvant chemotherapy after ablation and systemic therapy in early relapse. In this study, we investigated the possibility of pathological evaluation and gene mutation analysis using post-RFA liver tumor biopsy specimens.
METHODS
Animal experiments
A 15-day-old male mouse of C57BL/6 strain was injected intraperitoneally with diethylnitrosamine (Sigma) (25 mg/kg) and killed at 40 weeks of age. The developed liver tumor was resected and divided into two pieces: one for nontreated (unablated) tumor sequencing, the other for post-RFA tumor sequencing (Figure 1a). A 17G, cooled-tip electrode was inserted to the divided liver tumor sample for post-RFA sequencing and connected to a 500 kHz RF generator (Radionics), which produces 200 W at 50 Ω of impedance. Radiofrequency ablation was carried out until a rapid increase in impedance was observed. For pathological assessment, mouse livers were fixed with 4% paraformaldehyde and embedded in paraffin and tissue sections were subjected to H&E or silver-impregnated staining. All the experiments were carried out in accordance with protocols approved by the Animal Ethics Committee of the University of Tokyo. The study is reported in accordance with ARRIVE guidelines.
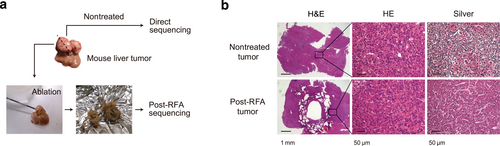
Radiofrequency ablation (RFA) for resected mouse liver tumor. (a) The resected mouse liver tumor was divided into two pieces: one for nontreated tumor sequencing, the other for post-RFA tumor sequencing. (b) Representative pathological images of the nontreated and post-RFA mouse liver tumors. H&E, hematoxylin and eosin.
Exome sequencing
Target selection and sequencing
Genomic DNA was extracted from mouse liver tumors and spleen (as a control) using a QIAamp DNA Mini Kit (Qiagen), sheared into approximately 200 bp fragments, and used to make a library for multiplexed paired-end sequencing with the SureSelectXT Reagent Kit (Agilent Technologies). The constructed library was hybridized to biotinylated cRNA oligonucleotide baits from the SureSelectXT Mouse All Exon Kit (Agilent Technologies) for target enrichment. Targeted sequences were purified by magnetic beads, amplified, and sequenced on an Illumina HiSeq 2500 platform in paired-end 101 bp configuration. Nucleotide sequence data reported are available in the DDBJ database under the accession numbers DRR396396–DRR396400.
Mapping and SNV/indel calling
Adapter sequences were removed by cutadapt (version 1.2.1). After quality control, reads were mapped to the reference mouse genome (mm10) using BWA (version 0.7.10). Mapping result was corrected using Picard (version 1.73) for removing duplicates and GATK (version 1.6-13) for local alignment and quality score recalibration. Single nucleotide variant and indel calls were undertaken with multisample calling using GATK and filtered to coordinates with VQSR passed and variant call quality score ≥30. Somatic SNV calls were performed by comparing tumor and normal pairs using SomaticSniper (version 1.0.2.3). Annotations of SNVs and indels were based on dbSNP138, CCDS (NCBI, Release Mm103), RefSeq (UCSC Genome Browser, November 2013). Variants were further filtered according to the following criteria: with predicted functions of frameshift, nonsense, read-through, missense, deletion, insertion, or insertion-deletion.
Human clinical samples and histopathological assessment
Percutaneous liver biopsy was undertaken on the same day as the liver tumor was treated with RFA, using a 15G needle with a biopsy specimen notch of 20 mm for background liver or a 20G needle for post-RFA liver tumor. Immediately after RFA, the tumor is difficult to detect due to the vapor, and a tumor biopsy was performed when the shape of the tumor was recognizable after 5–10 min (Figure S1). Liver biopsy specimens were assessed histologically by the expert pathologists (H.R. and M.T.). The fibrosis and inflammation in the background liver were assessed according to the METAVIR scoring system.10 Pathological evaluation was carried out based on the World Health Organization classification in 2019.11 Hepatocellular carcinoma was divided into three grades: well, moderately, and poorly differentiated. The study protocol was approved by the University of Tokyo Medical Research Center Ethics Committee (approval number 11839). All patients provided written informed consent. The methods were applied following the ethical guidelines of the Declaration of Helsinki.
DNA extraction of human liver samples and TERT mutations detection
DNA was extracted from human liver frozen samples obtained from post-RFA tumor using NucleoSpin tissue XS (Takara Bio) according to the manufacturer's protocol. The polymerase chain reaction primers were designed to amplify a 163 bp product containing hotspot mutation C228T in TERT promoter region (forward; 5′-GTCCTGCCCCTTCACCTT-3′; reverse, 5′-CAGCGCTGCCTGAAACTC-3′).
RESULTS
Histological findings of post-RFA liver tumor specimens from mouse
Radiofrequency ablation was performed on mouse liver tumor induced by diethylnitrosamine. The low-magnification image of the H&E stain of the mouse liver showed RFA needle penetration into the center of the specimen (Figure 1b, left). The high-magnification image showing H&E staining at the center of the ablated area was consistent with HCC, and there was no significant difference between the nontreated (unablated) and post-RFA tumors (Figure 1b, middle). However, silver-impregnated staining showed the disappearance of reticulin fibers in the post-RFA specimens compared with nontreated tumor, confirming that the RFA treatment had sufficiently disrupted the HCC structure (Figure 1b, right).
Exome sequencing using post-RFA mouse liver tumor
Whole exome sequencing was undertaken on the nontreated and post-RFA liver tumor samples from mouse. The DNA extracted from the post-RFA specimens formed a smear on electrophoresis, suggesting fragmentation, but the majority were approximately 20 kbp in length (Figure S2). We obtained high-quality nucleotide sequences covering 10.2 Gbp (121.3 × coverage) of the nontreated tumor genome and 10.9 Gbp (132.0 × coverage) of the post-RFA tumor genome (Table S1). We identified 2127 somatic mutations in the nontreated and 2395 somatic mutations in the post-RFA tumors, with approximately 80% in common with each other (Figure 2a, left). Furthermore, we identified 684 nonsynonymous somatic substitutions in the nontreated and 704 nonsynonymous somatic substitutions in the post-RFA tumors, with approximately 85% substitutions being significantly in common with each other (p < 0.001, hypergeometric probability test; Figure 2a, right). The average somatic mutation rate was comparable between the nontreated (41.33 nt/Mbp) and post-RFA (46.53 nt/Mbp) tumors (Table S1). There was no obvious change in the mutation site, with or without RFA (Figure 2b, Table 1). Small indels such as insertions and deletions of alleles and the patterns of base transitions were also detected comparably in the post-RFA sample (Figure 2b, Table 1). To determine the reliability of the mutational profiles, the overall distribution of mutant allele frequencies of total detected somatic alterations was examined; median frequencies were not significantly different between the nontreated and the post-RFA sample (0.270 [0.200–0.324] vs. 0.270 [0.195–0.324]; p = 0.24, t-test) (Figure 2c). We undertook Sanger sequencing to verify whether somatic mutations could be detected in the post-RFA samples. Allele changes in Sox2, Myc, and Casp1, showing high mutant allele frequencies in the nontreated tumor genome, were comparably detected in the post-RFA tumor genome (Figure 2d, Table 2). These results suggest that thermal denaturation by RFA had no significant effect on the detection of gene mutations.
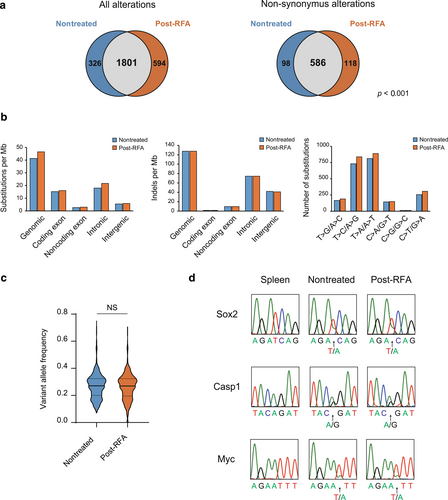
Results of whole genome sequencing using mouse liver tumors. (a) Number of somatic alterations in the nontreated and postradiofrequency ablation (RFA) tumors. Nonsynonymous alterations were significantly in common between nontreated and post-RFA tumors (p < 0.001, hypergeometric probability). (b) Prevalence of somatic substitutions, small indels, and patterns of base transitions in different genome regions. (c) Violin plots of variant allele frequency of somatic alterations detected in nontreated tumor and post-RFA tumor. The thick line inside represents the median, and the thin lines represent the first and third quartiles. (d) Electropherograms of the sequencing of indicated sites of Sox2, Myc, and Casp1 in spleen, nontreated tumor, and post-RFA tumor. NS, not significant.
| Type of changes | Tumor | Post-RFA |
|---|---|---|
| Substitutions | 2127 | 2395 |
| Coding | 781 | 823 |
| Nonsense | 38 | 41 |
| Missense | 551 | 583 |
| Synonymous | 188 | 195 |
| Unknown | 4 | 4 |
| Noncoding | 135 | 154 |
| UTR | 71 | 85 |
| Upstream | 22 | 26 |
| Downstream | 11 | 10 |
| ncRNA | 31 | 33 |
| Intronic | 927 | 1115 |
| Splice site | 26 | 23 |
| Other | 901 | 1092 |
| Intergenic | 282 | 301 |
| Unknown | 2 | 2 |
| Small indels | 6584 | 6584 |
| Coding | 59 | 60 |
| Frameshift | 29 | 30 |
| Insertion | 10 | 10 |
| Deletion | 13 | 13 |
| Insertion-deletion | 1 | 1 |
| Unknown | 6 | 6 |
| Noncoding | 528 | 531 |
| UTR | 231 | 239 |
| Upstream | 135 | 127 |
| Downstream | 38 | 37 |
| ncRNA | 124 | 128 |
| Intronic | 3838 | 3865 |
| Splice site | 32 | 34 |
| Other | 3806 | 3831 |
| Intergenic | 2159 | 2128 |
- Abbreviations: ncRNA, noncoding RNA; RFA, radiofrequency ablation; UTR, untranslated region.
| Gene | Chr. | Position | Allele change | Function | VAF | Depth | ||
|---|---|---|---|---|---|---|---|---|
| Tumor | Post-RFA | Tumor | Post-RFA | |||||
| Sox2 | 3 | 34,650,633 | T > A | Missense | 0.3162 | 0.2422 | 136 | 128 |
| Myc | 15 | 61,987,611 | T > A | Splice acceptor site | 0.2357 | 0.3946 | 140 | 147 |
| Casp1 | 9 | 5,302,996 | A > G | Missense | 0.3628 | 0.4608 | 113 | 102 |
- Abbreviations: Chr., chromosome; RFA, radiofrequency ablation; VAF, variant allele frequency.
Detection of gene mutation in post-RFA human liver tumors
Post-RFA human liver tumor specimens were used to detect mutations in the TERT promoter, the most prevalent genetic alteration in HCC.12 We focused on five cases in which the post-RFA tumor specimens had pathological evidence consistent with HCC (Figure 3a). The background profiles of these patients are presented in Table 3. In the case of HCC3, immunohistochemical analysis of glypican-3 was helpful for diagnosis of HCC. Approximately 0.5–1.0 μg DNA could be extracted from 1 mm3 of post-RFA tumor samples. TERT promoter mutations (C228T) were detected in HCC2 and HCC3, whereas no mutations were detected in nontumor areas (Figure 3b). HCC1 had no background liver samples.
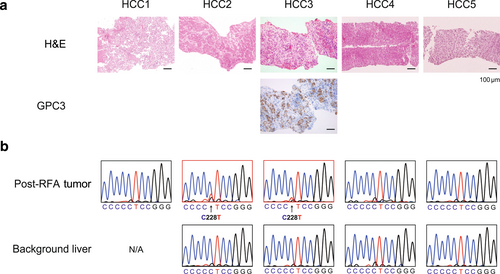
Detection of TERT promoter mutations using human postradiofrequency ablation (RFA) specimens. (a) Representative pathological findings of human post-RFA hepatocellular carcinoma (HCC). Tumor cells of case HCC3 showed positive for glypican-3 (GPC3). (b) Electropherograms of the sequencing of the TERT promoter in human liver biopsy samples. H&E, hematoxylin and eosin; N/A, not applicable.
| Case | Sex | Age (years) | Etiology | Size (cm) | Pathology | BG liver | Mutation |
|---|---|---|---|---|---|---|---|
| HCC1 | M | 67 | HCV | 4.9 | Well to moderately differentiated HCC | - | − |
| HCC2 | M | 62 | HCV | 3.1 | Well to moderately differentiated HCC | F4A1 | + |
| HCC3 | M | 64 | HCV | 2.9 | Well to moderately differentiated HCC (GPC3+) | F4A2 | + |
| HCC4 | M | 68 | HBV | 2.7 | Well differentiated HCC | F3A0-1 | − |
| HCC5 | F | 70 | AIH + PBC | 2.0 | Well to moderately differentiated HCC | F3-4A- | − |
- Abbreviations: AIH, autoimmune hepatitis; BG, background; F, female; GPC3, glypican3; HBV, hepatitis B virus; HCV, hepatitis C virus; M, male; PBC, primary biliary cholangitis.
Case report
We present a striking case in which the pathological evaluation of post-RFA tumor specimens was clinically useful. The patient was a 64-year-old man with a 3.6 cm liver tumor originating from chronic hepatitis B. The contrast pattern on CT images was atypical for HCC; the contrast effect on the edge of the tumor was enhanced in the portal phase and remained in the equilibrium phase (Figure 4a). Surgical treatment was suggested, but RFA was carried out at the patient's request. Pathological evaluation of the post-RFA biopsy specimen led to a diagnosis of adenocarcinoma (Figure 4b). The absence of malignant tumors in organs other than the liver suggested that the patient had ICC due to chronic hepatitis B.
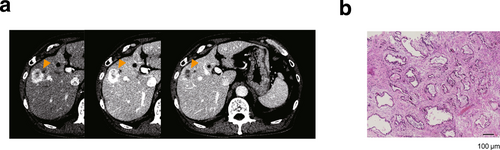
Case of a 64-year-old man with intrahepatic cholangiocarcinoma arising in chronic hepatitis B. (a) Radiological images of contrast-enhanced computed tomography. A hypervascular tumor 3.6 cm in diameter was seen in segment eight of the liver (arrowheads). Enhancement on the edge of the tumor at arterial and portal phase was seen, while contrast effect in the center of the tumor was poor. Contrast agent wash-out was seen at late phase. (b) Pathologically, tubular adenocarcinoma with desmoplastic reaction was observed, consistent with intrahepatic cholangiocarcinoma.
DISCUSSION
Tumor biopsy is essential for a definitive diagnosis of malignancy. However, due to the risk of neoplastic seeding, biopsy may not be performed prior to the treatment of liver tumors that are curable by surgery or localized therapy. In the case of hepatic resection, pathological evaluation is possible afterward; however, in the case of RFA, pathological evaluation is usually impossible, and the nature of the treated tumor will not be revealed. To the best of our knowledge, this is the first report to determine the clinical utility of post-RFA liver tumor samples and their application in gene mutation analysis.
In the early 2000s, when RFA was first introduced, various studies had focused on the pathological evaluation of tumors after RFA treatment.13-16 Experimentally, it has been shown that apoptotic changes spread over several days after RFA in the tissue surrounding the center of the ablated area where “coagulation necrosis” occurs.17 Evidence suggests that focal hyperthermia produces both direct and indirect tissue injury through different underlying processes.18 “Thermal fixation,” where the gross tissue architecture and microscopic details are preserved, and “apoptotic changes” associated with hyperthermia were considered as other forms of cell death, in addition to “coagulation necrosis” after RFA.19 As the cell damage after RFA spreads gradually over several days, there could exist areas immediately after the RFA where cellular degeneration has not yet started, and the pathological assessment of the tumor cells can be performed.
Society guidelines insist on the noninvasive diagnosis of HCC in cirrhotic patients and that tumor biopsies are only necessary if contrast-enhanced CT or magnetic resonance imaging does not show the typical hallmarks of HCC.20-22 In practice, however, even atypical HCC based on imaging analysis may be subjected to RFA without biopsy to avoid the risk of impairing curative treatment due to neoplastic seeding. We have previously shown that the risk of neoplastic seeding can be significantly reduced by performing liver tumor biopsies after RFA.9 It is worthwhile to challenge the pathological assessment of a post-RFA tumor biopsy, as it can avoid the risk of neoplastic seeding. In fact, we have encountered a patient with liver tumor derived from chronic hepatitis B, where the radiological imaging was atypical for HCC, and the diagnosis of adenocarcinoma could be made by the pathological assessment of post-RFA biopsy. The patient had ICC, which is known to occasionally occur in chronic liver disease.23, 24 Furthermore, in some cases, immunostaining might be possible in post-RFA specimens, suggesting that immunohistochemical analyses such as glypican-3 or alpha-fetoprotein could be used to diagnose HCC, even when pathological evaluation is difficult due to thermal degeneration. However, it should be noted that the possibility of immunostaining in post-RFA specimens was not investigated in many cases, and there may naturally be cases where immunostaining is not possible due to degeneration caused by ablation.
To investigate the feasibility of analyzing genetic mutations using post-RFA tissue, we undertook experiments using mouse liver tumors. Notably, the detected nonsynonymous mutations were consistent between nontreated and post-RFA tumors, suggesting that genome sequencing can be performed with comparable accuracy even after RFA. However, this experiment had some limitations. The nontreated and post-RFA sites were different lesions of the same tumor and not strictly post- and pre-RFA of the same sample. Moreover, the detected genetic variants were not completely identical, which could be due to tumor heterogeneity, but the effect of RFA on the sensitivity of sequencing cannot be completely ruled out. The number of SNVs detected in mouse liver tumors showed an extremely high tumor mutation burden (approximately 40 nt/Mbp). According to the previous report, diethylnitrosamine-induced liver tumors have indeed a high mutation burden of approximately 20 nt/Mbp, compared to tumors in transgenic mice.25 This could be because diethylnitrosamine induces significant DNA damage in juvenile mice. In the present study, larger tumors were selected for easier ablation, which may have resulted in an even higher mutation burden. For the human liver biopsy specimens, approximately 0.5 μg DNA could be extracted from 1 mm3 of post-RFA liver tumor specimen, which was further used to detect TERT promoter mutations.
The use of cell-free DNA or circulating tumor cells, known as “liquid biopsy,” has evolved as a noninvasive method for undertaking tumor genome analysis.26, 27 This method requires only blood sample and is less burdensome for the patient; however, it contains a large proportion of blood cell-derived materials and is still at infancy in detecting tumor-derived genetic mutations. In contrast, post-RFA tumor biopsy is closer to clinical application because of its simplicity and ability to sequence the tumor-derived genome, which is abundant in the specimen.
There are, however, some limitations to this method of using post-RFA tumor specimens. First, the post-RFA samples can only be used for mutation analysis, because only DNA can be extracted, which is resistant to thermal denaturation. Because the RNA is degraded, gene expression analysis is not possible. Second, DNA from post-RFA specimens can be fragmented depending on the degree of ablation, and therefore, it is likely that targeted deep sequencing or other sophisticated techniques will be required for comprehensive genetic mutation analysis, and further studies are needed in this regard. Third, as the biopsy is performed after RFA, there is a possibility that the biopsy needle will not reach the tumor due to technical problems. Finally, pathological evaluation is not always possible due to the degenerative effects of RFA.
Neoplastic seeding due to liver tumor biopsy is an infrequent but serious complication with a significant impact on prognosis. Post-RFA tumor biopsy is a useful and safe method for obtaining tumor samples that can be used for gene mutation analysis and pathological evaluation, which will contribute to the future HCC treatment where precision medicine based on genetic tumor mutations is expected.
ACKNOWLEDGMENTS
We would like to thank Editage for English language editing. This research was supported by Grants-in-Aid for Scientific Research 21K15989 (T.N.), 21H02892 (H.N.), and 20K08352 (R.T.), MSD Life Science Foundation, The Naito Foundation, Life Science Foundation of Japan, The Cell Science Research Foundation (H.N.), the Research Program on Hepatitis from Japan Agency for Medical Research and Development (AMED) under Grant Number JP21fk0210090 (H.N.), JP21fk0210059 (H.N.), JP22fk0210115 (H.N.), and JP22fk0210066 (R.T.).
CONFLICT OF INTEREST STATEMENT
The authors have no relevant financial or nonfinancial interests to disclose. Hayato Nakagawa and Ryosuke Tateishi are Editorial Board members of Hepatology Research and co-authors of this article. To minimize bias, they were excluded from all editorial decision-making related to the acceptance of this article for publication.
ETHICS STATEMENT
Approval of the research protocol: The clinical study protocol was approved by the University of Tokyo Medical Research Center Ethics Committee (approval number 11839).
Informed consent: All patients provided written informed consent.
Registry and the registration no. of the study/trial: N/A.
Animal studies: All the animal experiments were performed in accordance with protocols approved by the Animal Ethics Committee of the University of Tokyo.
Research involving recombinant DNA: N/A.
Open Research
DATA AVAILABILITY STATEMENT
The datasets generated and/or analyzed during the current study are available from the corresponding authors upon reasonable request. Nucleotide sequence data reported are available in the DDBJ Sequenced Read Archive under the accession numbers DRR396396–DRR396400. The data have been deposited with links to BioProject accession number PRJDB14099 in the DDBJ BioProject database (https://ddbj.nig.ac.jp/resource/bioproject/PRJDB14099).




