Different immortalized keratinocyte cell lines display distinct capabilities to differentiate and reconstitute an epidermis in vitro
Abstract
Dermatological research relies on the availability of suitable models that most accurately reflect the in vivo situation. Primary keratinocytes obtained from skin reduction surgeries are not only limited by availability but have a short lifespan and show donor-specific variations, which hamper the understanding of general mechanisms. The spontaneously immortalized keratinocyte cell line HaCaT displays chromosomal aberrations and is known to differentiate in an abnormal manner. To overcome these issues, we validated different engineered immortalized cell lines created from primary human keratinocytes (NHK) as model systems to study epidermal function. Cell lines either immortalized by the expression of SV40 large T antigen and hTERT (NHK-SV/TERT) or by transduction with HPV E6/E7 (NHK-E6/E7) were analysed for their growth and differentiation behaviour using 2D and 3D culture systems and compared to primary keratinocytes. Both cell lines displayed a robust proliferative behaviour but were still sensitive to contact inhibition. NHK-E6/E7 could be driven into differentiation by Ca2+ switch, while NHK-SV/TERT needed withdrawal from any proliferative signal to initiate a delayed onset of differentiation. In 3D epidermal models both cell lines were able to reconstitute a stratified epidermis and functional epidermal barrier. However, only NHK-E6/E7 showed a degree of epidermal maturation and stratification that was comparable to primary keratinocytes.
1 INTRODUCTION
The epidermis forms the outermost barrier between the human body and the environment. It consists of up to 95% of keratinocytes1 and constantly regenerates itself.2 This is sustained by epidermal stem cells constantly dividing into transient amplifying cells, that in turn initiate an organized program of differentiation and migration to the upper layers.3 During this process, cells change their transcriptional program and eventually develop into the defined layers of the epidermis, each characterized by specific structural proteins.3, 4 At the end of this differentiation process keratinocytes have matured into corneocytes that form the protective outer corneal layer. In inflammatory skin diseases this tightly regulated homeostasis is often disturbed leading to impaired barrier function. Thus, to better understand the epidermal physiology, investigate therapeutic targets and evaluate novel topical treatments pre-clinically, in vitro models are required, that resemble the in vivo situation in the epidermis most accurately.
The differentiation program can be mimicked in vitro, by treating 2D monolayer cell cultures with elevated Ca2+ concentrations5, 6 or post-confluent growth.7, 8 For an even better understanding of the epidermal barrier formation and function, 3D models are generated by cultivating confluent keratinocytes at the air-liquid interface to initiate differentiation, giving rise to a fully stratified and functional epidermis.9 These techniques can then be combined with inflammatory mediators such as Th1 or Th2 cytokines, gene editing methods like CRISPR/Cas9, or the use of patient derived cells to generate disease models for common dermatoses such as psoriasis10, 11 or atopic dermatitis.12, 13
All these models rely on the use of normal human epidermal keratinocytes (NHK) that are isolated from donor skin obtained from plastic surgery or circumcisions. However, these cells are limited by availability and display donor-specific variations in terms of (epi) genetic background, age or body site that hamper the understanding of general principles. In addition, primary cells have a short life span, which makes gene editing difficult as isolation of clonal cell lines is nearly impossible.14 For these reasons, many researchers have used the spontaneously immortalized keratinocyte cell line called HaCaT,15 which helped to understand many characteristics of keratinocytes. However, HaCaT cells do not adequately reflect the epidermal differentiation pattern, as they poorly respond to Ca2+ and express envelope proteins like filaggrin and loricrin abnormally.16, 17 In vitro reconstituted human epidermis from HaCaT cells is not well stratified and completely lacks a stratum corneum, which is associated with decreased barrier function.18, 19 Although HaCaT cells are widely used for genome-editing due to the fact that single cell clones can be isolated and easily propagated,20 they display chromosomal aberrations and show aneuploidy.15
To overcome these issues, several immortalization approaches have been developed including ectopic expression of human telomerase (hTERT),21, 22 the cell cycle checkpoint factor Cdk4 and hTERT,23 the SV40 (simian virus 40) large T antigen,24 the HPV (human papillomavirus) E6 and E7 oncogenes25 or treatment with a Rho kinase inhibitor.26 Most of these cell lines retain characteristics of primary keratinocytes, including epidermal adhesion,27 barrier function18 and growth in 3D epidermal28 or full thickness skin equivalents.21, 26, 29 However, the degree of similarity with primary keratinocytes is divergent especially with regard to their ability to differentiate and reconstitute a stratified epidermis and a comprehensive analysis comparing cell lines immortalized by different approaches is lacking. Thus, we aimed at comparing the commercially available cell line NHK-SV/TERT (NHEK/SV-TERT3-5, Evercyte GmbH), generated from adult abdominal keratinocytes by overexpression of SV40 large T antigen and hTERT,30, 31 and our own cell line NHK-E6/E732 generated from juvenile keratinocytes after retroviral transfection with HPV oncogenes E6/E7. These cell lines are evaluated regarding their growth and differentiation capacities and suitability as in vitro models in comparison to primary NHK and HaCaT cells.
We could show that both cell lines can be alternatives to primary cells regarding their potential to differentiate; however, NHK-E6/E7 reconstitute an epidermis in 3D in vitro models that closer resembles the native epidermis. Hence, this cell line is well suitable for studies on epidermal biology, the generation of inflammatory disease model and pre-clinical testing of novel therapeutic approaches.
2 METHODS
2.1 Antibodies
Involucrin (ab20202 and ab181980) and keratin 10 (ab76318) antibodies were obtained from Abcam. Keratin 14 antibody (MCA890) was from BioRad. PCNA (MAB424R) was from Merck and claudin 1 (51-9000) antibody was purchased from Invitrogen. β-actin (A1978) and α-tubulin (T9026) antibodies were from Sigma and filaggrin (FLG) (905804) from BioLegend. Ki-67 (SP6) antibody was obtained from DCS. P-EGFR Y1068 (#3777), P-AKT S473 (#4060), P-ERK 1/2 T202/Y204 (#4370) and P-S6 S235/6 (#2211) were from Cell Signaling Technology.
2.2 Cells and cell culture
HaCaT cells were obtained from Cell Lines Services (CLS) and cultured in DMEM (Invitrogen), 10% FCS (Biochrom), 1% Pen/Strep solution (Invitrogen). Primary NHK (normal human keratinocytes) cells were isolated from human juvenile foreskin or various adult body regions after skin reduction surgery. Keratinocytes immortalized by the expression of SV40 large T antigen and hTERT (NHK-SV/TERT3-5) were purchased from Evercyte GmbH. The proprietary cell line NHK-E6/E7 was generated by lentiviral transduction of juvenile NHKs (purchased from PromoCell) using HPV16-E6/E7 human papillomavirus oncogenes (BRAIN Biotech AG).32 The latter three cell types were cultured in KGM-2 (PromoCell).
2.3 Immunofluorescence
2 × 105 cells were seeded onto glass coverslips in 12-well format. The next day, cells were fixed and permeabilized with ice cold methanol. Cells were blocked with 10% normal goat serum (Vector Laboratories) in PBS for 1 h at room temperature (RT). 10 μg/mL anti-β-actin-antibody was used for staining in 10% normal goat serum for 1 h at RT. After washing, antibody was detected with AlexaFluor488 labelled anti-mouse antibody (A11029, Life Technologies) for 1 h at RT. Cells were washed and stained for 5 min at RT with NucBlue Fixed Cell Stain Ready Probes Reagent (Invitrogen) and mounted with Fluoromount-G (Invitrogen). Images were acquired by using a Nikon Eclipse Ci microscope.
2.4 Transfection and flow cytometry
1.6 × 105 cells were seeded in 6-well plates. The next day, cells were transfected with 1 μg pcDNA3.1-acGFP (gift from Oskar Laur (Addgene plasmid #128047; http://n2t.net/addgene:128047; RRID: Addgene_128047)) using 3 μL Lipofectamine 3000 (Thermo Fisher) in 200 μL OptiMEM (Thermo Fisher) according to the manufacturer's instructions. Forty-eight hours post-transfection, cells were detached using Accutase (HaCaT pre-treated with 0.2% EDTA) and washed three times with Stain Buffer (BSA) (BD Biosciences). Cells were resuspended in Stain Buffer (BSA) and green fluorescence measured using the FACSCalibur. Gates were set according to untreated cells. Analysis was performed using the FlowJo Software (Version 10.8.1).
2.5 Proliferation assay
Cell proliferation was quantified by a colorimetric WST-1 assay (Roche) according to the manufacturer's instruction. Briefly, 7 × 103 cells were seeded in 96-well plates and the first time point (0 h) was assessed on the same day after cell attachment by the addition of WST-1. WST-1 metabolization was measured after another 4 h. For the following time points the procedure was repeated every 24 h.
2.6 Reverse transcription quantitative real-time PCR (RT-qPCR)
Total RNA was isolated using NucleoSpin RNA isolation kit (Macherey&Nagel), transcribed with the High-Capacity RNA-to-cDNA™ Kit (Thermo Fisher) and subjected to RT-qPCR using predesigned TaqMan® Gene Expression Assay probes (Thermo Fisher) on Step One Plus PCR System (Applied Biosystems) mRNA expression was normalized to RPLP0 and relative changes in the respective mRNA were quantified by the 2−ΔΔCt method.
2.7 Transcriptome data analysis
RNA was isolated using NucleoSpin RNA isolation kit (Macherey&Nagel). The TruSeq RNA Library Prep Kit v2 from Illumina was used with an input of 500 ng total RNA to prepare the library. Reads were trimmed with Trimmomatic 0.39,33 aligned to the hg19 reference genome using Hisat2 (2.2.1).34 Output files were sorted using SAMtools (1.10) and reads counted with HTSeq (1.99.2). Counts were normalized and differentially expressed genes identified using DESeq2 (1.32.0).35
2.8 Western blot analysis
Cells were lysed in RIPA lysis buffer (Cell Signaling Technology), while epidermal models were lysed by sonication in SDS lysis buffer (55 mM Tris pH 6.8; 2.2% SDS; 11% Glycerol). Protein amounts were normalized, subjected to SDS–PAGE and blotted onto PVDF membranes. After blocking in 5% milk/TBS-T, membranes were probed with the indicated antibodies and visualized with HRP-conjugated secondary antibodies using ECL Substrate (BioRad).
2.9 In vitro reconstructed human epidermis models
5 × 105 keratinocytes were seeded onto 12-well 0.4 μm pore size ThinCerts with PET-membranes (Greiner Bio-One GmbH, Frickenhausen, Germany) submerged in CnT-Prime Medium (CELLnTEC, Bern, Switzerland). After 3 days, medium was changed to differentiation medium (3 parts CnT-Prime 3D Barrier Medium (CELLnTEC)/2 parts DMEM). On the next day inserts were transferred to deep well plates and lifted to the air-liquid interface to induce keratinocyte maturation indicated by keratinization and stratification. Medium was changed every other day and models were harvested after 10 days. The membrane was cut out of the insert and divided into three parts to be used for protein and RNA extraction as well as histological analysis. For the latter, models were fixed in formalin and paraffin embedded. 4 μm sections were haematoxylin and eosin (HE) stained or used for immunohistochemical staining.
2.10 Epidermal barrier assessment
Transepithelial electrical resistance (TEER) measurements—To assess the water-soluble ion permeability of the epidermis, inserts were transferred to a regular 12-well plate with 500 μL medium on top and 1 mL within the well. Electrical resistance was measured at four different sites using an EVOM2 voltohmmeter (World Precision Instruments, Friedberg, Germany) with an STX2 electrode. Readings from a blank insert were subtracted and unit area resistance (Ωcm2) was calculated by multiplying resistance readings by the effective surface area.
Lucifer yellow dye and biotin penetration—To assess the outside-in barrier function, 1 mM Lucifer yellow (Sigma-Aldrich) was applied on top of the model for 60 min at 37°C. Subsequently, the inside-out barrier function was determined by turning the ThinCert insert upside down and applying 1.67 mg/mL EZ-Link™ Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific) on the bottom of the insert membrane for 60 min at RT. Models were fixed in formalin and paraffin embedded. 4 μm sections were deparaffinized and stained with 2 μg/mL Alexa Fluor 594 streptavidin (Thermo Fisher Scientific) for 30 min. Sections were mounted in VECTAshield, containing DAPI (Biozol).
2.11 Immunohistochemistry
Paraffin sections were processed routinely and stained over night with primary antibody or concentration adjusted isotype control antibody. Visualization was performed with Histofine Simple Stain AP Multi (Nichirei Bioscience) and nuclei were stained with haematoxylin. Images were acquired by using a Nikon Eclipse Ci microscope.
2.12 Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.5.0 using a 2-way ANOVA test and Fisher's LSD test for multiple comparison. p < 0.05 was considered statistically significant and were indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
3 RESULTS
3.1 Immortalized cell lines show a proliferative behaviour similar to primary keratinocytes and are inhibited by contact inhibition
We validated two immortalized keratinocyte cell lines that were generated either by the expression of SV40 large T antigen and hTERT (NHK-SV/TERT) or HPV E6/E7 oncogenes (NHK-E6/E7). Using β-actin staining we visualized the morphology of the different cells. In contrast to HaCaT cells, NHK-E6/E7 and NHK-SV/TERT show a cobblestone like shape similar to NHK. NHK-SV/TERT, however, appear smaller than primary NHK (Figure 1A).
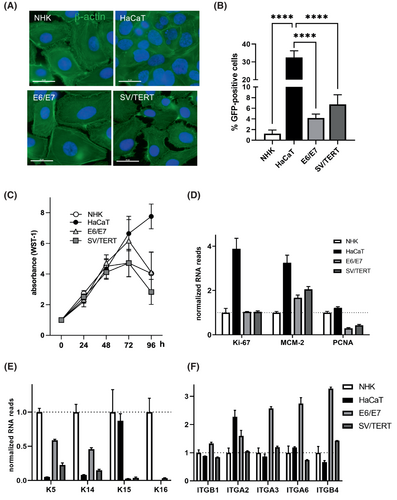
As efficient transfection is needed for different experimental approaches, we assessed the cells' capacity to be targeted by lipofection. As reported before HaCaT cells display transfections efficiencies up to 30%, while only 1% of primary NHK cells can be transfected using Lipofectamine 3000. In contrast, immortalized cell lines show around 4 (NHK-E6/E7) or six times (NHK-SV/TERT) higher transfection efficiencies but are still far below the efficiencies that can be achieved in HaCaT cells (Figure 1B). Both immortalized cell lines show indefinite growth up to over 50 passages and display growth rates comparable to primary cells as measured by WST-1 metabolization (Figure 1C). NHK-E6/E7 and NHK-SV/TERT show contact inhibition similar to NHK while HaCaT continue to grow (Figure 1C). Moreover, we analysed the proliferative profile by RNA sequencing. This reveals high expression levels of proliferation (Ki-67) and replication (MCM-2, PCNA) marker genes in HaCaT cells, while the immortalized cell lines show similar expression of Ki-67 as NHKs (Figure 1D). Markers that are specifically indicative of the basal, proliferative compartment of the epidermis such as keratin 5 (KRT5) and 14 (KRT14), are very little expressed in HaCaT cells, while they are more abundant in NHK-E6/E7 than in NHK-SV/TERT. Interestingly, the basal keratin 15 (KRT15) almost reaches the expression levels of NHK in HaCaT cells, while being little expressed in the immortalized cell lines. Conversely, keratin 16 (KRT16) can only be detected in NHK (Figure 1E). In healthy epidermis, keratinocytes only proliferate in the basal layer and cell attachment to the basal membrane through integrin-ligand binding is a crucial regulatory mechanism.36 Thus expression of the integrin β1 subunit (ITGB1) especially when forming a heterodimer with α2 (ITGA2) and α3 (ITGA3) as well as the α6β4 (ITGA6/ITGB4) heterodimer can be correlated with the proliferative potential of keratinocytes.37 Thus, we investigated the expression of these subunits, and found that compared to NHK, HaCaT cells strongly express integrin α2 (ITGA2), while the expression of all other investigated integrin subunits is especially high in NHK-E6/E7. NHK-SV/TERT show only moderate elevation of integrin subunits (Figure 1F).
3.2 NHK-E6/E7 can be differentiated by Ca2+ switch, while NHK-SV/TERT need withdrawal of any proliferative stimulus to properly commit to differentiation
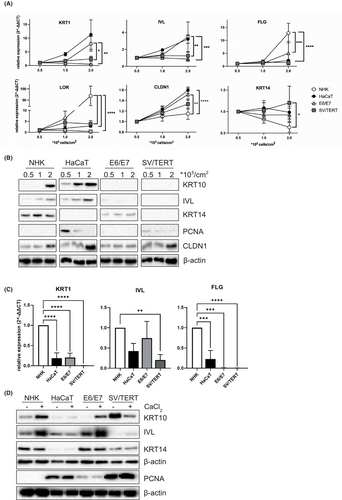
We analysed the capacity of the cell lines to differentiate in 2D cell culture by post-confluent growth. Seeding ascending cell numbers, NHK and HaCaT show increased expression on the RNA and protein level of keratin (KRT) 1 and 10, involucrin (IVL) and the tight junction marker claudin 1 (CLDN1) (Figure 2A,B), while only NHK are able to strongly express the late differentiation markers such as filaggrin (FLG) and loricrin (LOR) (Figure 2A). At the same time, cells downregulate proliferation markers such as keratin 14 or PCNA (Figure 2A,B). In contrast, in both immortalized cell lines the expression of all markers remains unregulated (Figure 2A,B), only the expression of claudin 1 could be induced by post-confluent growth in NHK-SV/TERT cells (Figure 2A,B). Next, we tried to promote differentiation using a Ca2+ switch. NHK cells can be very well induced to differentiate under these conditions, as shown by the expression of keratin 10 and involucrin (Figure 2D), thus for RNA expression analyses cell lines are compared relative to values obtained from NHK (Figure 2C). In comparison with NHK, immortalized cell lines show little to no expression of differentiation markers, only NHK-E6/E7 can be forced to express comparable levels of involucrin protein and RNA and keratin10 protein (Figure 2C,D). No expression of the late differentiation marker filaggrin could be detected (Figure 2C). Interestingly, NHK-SV/TERT express high levels of keratin 10 when kept in the differentiation medium in the absence of Ca2+, that are reduced when an additional Ca2+ stimulus is given (Figure 2D). Hence, these conditions are not suitable to induce differentiation in NHK-SV/TERT. In search of a possible reason for the cell lines' reduced ability to commit to differentiation, we analysed the RNAseq data for the expression of genes with a known role in controlling keratinocyte proliferation and found higher expression of genes of the EGFR/AKT (EGFR, GRB2, CCND1/Cyclin D1, FOXO3), MAPK (NRAS, KRAS and MAPK1/ERK2) and mTORC1 (RPTOR/Raptor, S6KA2/RSK3) signalling pathway in the immortalized cell lines compared to NHK. In contrast the phosphatase PTEN, that negatively regulates PI3K/PKB signalling38 is significantly downregulated in NHK-E6/E7 and NHK-SV/TERT (Figure 3A). Thus, hyperactivation of these pathways might prevent the initiation of differentiation. As these signalling cascades are very responsive to different growth factors and mitogens, we investigated whether withdrawal of any supplement would allow the cells to initiate differentiation and maturation. We could show that signalling molecules of these pathways such as EGFR, PKB, ERK1/2 and the ribosomal protein S6 are hyperphosphorylated especially in NHK-SV/TERT and only starvation from growth factors reduces activation efficiently (Figure 3B). Consequently, using a medium devoid of any supplements during differentiation diminished the expression of the proliferative markers PCNA and keratin 14 and allows induction of differentiation by Ca2+ as measured by the expression of keratin 10 and filaggrin, especially in NHK-SV/TERT cells (Figure 3C,D). However, it must be noted, that the increase in mRNA expression in NHK-SV/TERT under starved conditions seems impressive, when depicted here as fold induction relative to the untreated control (Figure 3C). Nevertheless, the total expression of the differentiation markers is still lower compared to the other cell types (Figure 3D). Interestingly, in the NHK-E6/E7 cell line growth factor withdrawal has only a small effect on the expression of keratin 1/10 while the expression of claudin 1 increased greatly under starvation condition in all cell types, even without any additional effect of Ca2+ (Figure 3D).
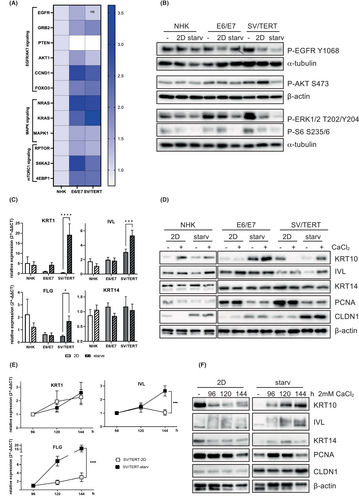
Since the degree of differentiation in NHK-SV/TERT cells is still not satisfactory, we speculate that the switch from proliferation to differentiation might take longer. Indeed, the expression of differentiation markers increased continuously for up to 6 days after Ca2+ stimulation, especially under starved conditions (Figure 3E,F), while in the NHK-E6/E7 cell line no additional increase in differentiation markers could be detected (data not shown). Thus, both immortalized cell lines can be induced to differentiate with NHK-SV/TERT cells needing growth factor withdrawal and extended Ca2+ stimulation.
3.3 NHK-E6/E7 reconstitute an epidermis comparable to primary keratinocytes
Next, we analysed the potential of the immortalized cell lines to be used in 3D reconstituted epidermal models. As expected, NHK cells are able to reconstitute a well stratified epidermis, covered by a corneal layer, while HaCaT cells form an epidermis devoid of a stratum granulosum and corneum (Figure 4A). In contrast, both immortal cell lines generated a stratified epidermis consisting of all four epidermal layers (Figure 4A), which was thinner than the epidermis grown from NHKs (Figure 4B). These models displayed a functional epidermal barrier as shown by the exclusion of lucifer yellow (LY) that was comparable to primary cells, while in HaCaT models the dye cannot be retained but completely passages through the epidermis (Figure 4C). As an additional measure of barrier function, transepithelial electrical resistance (TEER) was gaged, showing that the values achieved in the immortalized cell lines are lower, but still superior to HaCaT cells (Figure 4D). Claudin 1, as a major component of tight junctions and thus also contributing to barrier function in the stratum spinosum is expressed in all cell lines but displays abnormal distribution throughout the whole epidermis in HaCaT. As the models generated from the immortalized cell lines are a lot thinner, proper localization is difficult to estimate, but in the NHK-E6/E7 models claudin 1 localization at the cell membrane of cells in the stratum granulosum can be assumed (Figure 4E). Next, we assayed the degree of differentiation that can be achieved in these models by immunohistological staining, RT-qPCR and western blot analysis. The NHK-E6/E7 cell line shows expression levels of keratin 10, involucrin and filaggrin that are comparable or even superior to NHKs. In contrast, HaCaT and NHK-SV/TERT display aberrant expression and distribution of these markers (Figure 4F–H). While in NHK and NHK-E6/E7 proliferative cells, as indicated by Ki-67 positive nuclei, were confined to the basal layer, HaCaT and especially NHK-SV/TERT showed Ki-67 positive nuclei in suprabasal layers (Figure 4I). Keratin 14 as another marker of dividing keratinocytes is expressed in the basal epidermal layer in models from primary cells, while being aberrantly distributed in epidermis reconstituted from the cell lines (Figure 4I). Thus, both immortalized cell lines can reconstitute an epidermal barrier, but only NHK-E6/E7 cells show features of stratification and differentiation that are comparable to primary cells.
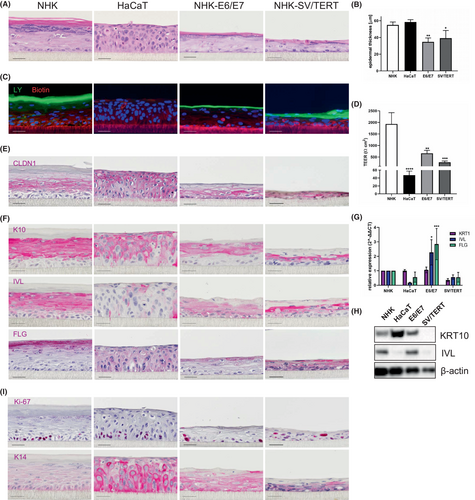
In summary, NHK-E6/E7 show a differentiation behaviour comparable to primary cells, while the highly proliferative NHK-SV/TERT cells need withdrawal of any proliferative stimulus to commit to a delayed onset of differentiation. In 3D epidermal models both cell lines were able to reconstitute an epidermis that was thinner than that formed by primary cells, but with a tight and functional barrier. However, only NHK-E6/E7 display features of keratinocyte maturation and stratification.
4 DISCUSSION
Dermatological research depends on adequate models to study epidermal physiology and pathomechanisms underlying different dermatological conditions. Most approaches rely on the availability of primary donor cells which suffer from issues such as limited availability, donor-specific variations and finite lifespan. In accordance with others, we show here that the spontaneously immortalized cell line HaCaT is not an optimal model as the cells show aberrant expression of differentiation marker19 and do not form a proper epidermal barrier.18 Thus, different approaches were undertaken to engineer immortalized keratinocyte lines that circumvent these difficulties. Normal human cells are doomed for senescence as telomeres shorten during each replicative cycle. Thus, immortalization of cells was tried to achieve by stabilization of telomeres through expression of hTERT.39 However, hTERT expression alone seems to be insufficient to evade senescence in epithelial cells such as keratinocytes, which rather require additional manipulation of the cell cycle regulators pRB or p53.21, 40 Thus, the first cell line we evaluated (NHK-SV/TERT) was immortalized by a two-step process comprising ectopic expression of SV40 T antigen and hTERT31 and displayed a delayed onset of differentiation and reconstituted thinner, less differentiated epidermal models. Although NHK-SV/TERT exhibited cellular growth comparable to NHK, proliferative markers were not downregulated, when differentiation was initiated and were also detected outside the basal layer in epidermis equivalents. This might be not only due to upregulation of growth and proliferation mediating pathways such as the EGFR/AKT, MAPK and mTORC1 cascade, but also due to SV40 T antigen overexpression, which binds to the cell cycle regulators pRB as well as p53 to promote DNA replication and complements hTERT in bypassing cellular senescence.41 Hence, NHK-SV/TERT need depletion from any proliferative stimulus to withdraw from the cell cycle and commit to differentiation adequately. Wagner et al., showed that in SV40 and hTERT immortalized cells, SV40 is expressed in basal cells of 3D models, but was downregulated in suprabasal keratinocytes, allowing ordered differentiation.31 However, in our reconstituted epidermal models, proliferative (Ki-67+) cells could be still detected in upper epidermal layers. While we used epidermis-only models, previous work achieved epidermal differentiation in full-thickness models.30, 31 Thus, factors from the underlying fibroblast containing collagen matrix that reconstitute the basal lamina might retain those highly proliferative cells in the basal compartment, allowing only cells fully committed to differentiation to form suprabasal layers.42 Although the expression of differentiation markers was reduced in our models and stratification was less pronounced, the models displayed a stratum granulosum and stratum corneum contributing to a fully functional epidermal barrier.
In contrast to our results, a cell line (N/TERT) that was also obtained by expression of hTERT21 showed differentiation and stratification comparable to primary keratinocytes.18, 21, 28, 29, 43 While N/TERT are deficient for p16INK thus leading to p53 inhibition,21 the SV40 T antigen in NHK-SV/TERT deregulates the p53 as well as the pRB pathway.41, 44 Consequently, it might be easier for N/TERT to withdraw from the cell cycle and commit to differentiation.
An alternative strategy to immortalize cells is overexpression of HPV16 E6/E7 genes. The human papilloma virus 16 contributes to the development of most cervical cancers by mediating transformation of keratinocytes and abrogating gene expression associated with epithelial barrier, such as intercellular junctions, and cell adhesion.45, 46 Especially the viral oncoproteins E6 and E7 mediate oncogenesis by interfering with the pRB and p53 pathways47 and promote proliferation while altering epidermal differentiation.48 Transducing organotypic skin cultures with HPV16 led to dysplasia, disturbed differentiation and Ki-67-positive cells in upper layers.49, 50 However, the epidermis generated from our NHK-E6/E7 cell line was satisfactorily differentiated and contained proliferative cells only in the basal layer, with proliferation markers such as PCNA or Ki-67 not being significantly upregulated. A few approaches were undertaken to use the E6/E7 oncogenes to generate immortal skin keratinocyte cell lines in vitro. While some of the generated cell lines were resistant to differentiation stimuli,51 others were able to express differentiation markers, but reconstituted an incompletely stratified epidermis.25 In contrast, our NHK-E6/E7 cell line performed much better by differentiating well in monolayer culture and reconstituting a well stratified epidermis very similar to primary keratinocytes. This might be due to different integration sites of the E6/E7 genes after lentiviral transduction. During keratinocyte differentiation nuclear remodelling and epigenetic modifications occur.52, 53 Hence, the E6/E7 oncogenes might be only expressed in the proliferative state, but are silenced during differentiation, preventing interference with the keratinocyte maturation process. Thus, our cell line immortalized with HPV E6/E7 genes can serve as a suitable substitute for primary cells to investigate epidermal maturation.
In conclusion, researchers should select the cellular model thoroughly depending on the epidermal mechanisms to be investigated. While the NHK-SV/TERT cell line can be very well used for studies examining epidermal barrier function, for approaches focusing on epidermal differentiation and stratification, full-thickness models including a dermal compartment should be preferred. In contrast the NHK-E6/E7 cell line is a versatile model, displaying good differentiation behaviour in 2D assays and reconstitutes a stratified and well functional epidermis in 3D assays even in the absence of a dermal compartment.
AUTHOR CONTRIBUTIONS
Magdalena Jahn, Torsten Fauth and Claudia Buerger designed the study with help of Roland Kaufmann; Robin Back, Magdalena Jahn, Sandra Diehl and Victoria Lang performed the experiments. Magdalena Jahn, Torsten Fauth and Claudia Buerger analysed the data; wrote the manuscript.
ACKNOWLEDGEMENTS
This work was funded by the Deutsche Forschungsgemeinschaft (DFG-German Research Foundation) BU1840/5-3, UfIB/NatLifE2020 German Federal Ministry of Education and Research 031B0716 as well as the Rolf. M. Schwiete Foundation (Germany). Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST STATEMENT
TF and RB are employees of BRAIN Biotech AG. RB owns shares of BRAIN Biotech AG. TF owns share options of BRAIN Biotech AG. All other authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.