GINS complex subunit 4, a prognostic biomarker and reversely mediated by Krüppel-like factor 4, promotes the growth of colorectal cancer
Abstract
GINS complex subunit 4 (GINS4) is essential for DNA replication initiation and elongation in the G1/S phase cell cycle in eukaryotes, however, its functional roles and molecular mechanisms remain unclear in many aspects. Our study was designed to investigate the clinical significance, biological function, and molecular mechanism of GINS4 in colorectal cancer (CRC). First, we confirmed that GINS4 expression was significantly overexpressed in CRC cells and tissues. The immunohistochemical results in tissue microarray from 106 CRC patients showed that a high level of GINS4 expression was positively correlated with advanced T stage, higher tumor TNM stage, and poor differentiation. The results from univariate and multivariate survival analysis models based on 106 CRC patients revealed that GINS4 might serve as an independent prognostic indicator for overall survival and disease-free survival of CRC patients. Moreover, downregulated GINS4 can inhibit growth and the cell cycle and accelerate cell apoptosis progression in vitro as well as inhibit tumorigenesis in vivo. Besides, our results also indicated that Krüppel-like factor 4 (KLF4) can negatively regulate GINS4 expression at the transcriptional level and the KLF/GINS4 pathway might play a vital role in the growth and prognosis of CRC.
Abbreviations
-
- CI
-
- confidence interval
-
- CRC
-
- colorectal cancer
-
- DFS
-
- disease-free survival
-
- EMT
-
- epithelial-mesenchymal transition
-
- GINS4
-
- GINS complex subunit 4
-
- HR
-
- hazard ratio
-
- IHC
-
- immunohistochemical
-
- KLF4
-
- Krüppel-like factor 4
-
- miR
-
- microRNA
-
- NDRG2
-
- N-Myc downstream-regulated gene 2
-
- OS
-
- overall survival
-
- PE
-
- phycoerythrin
-
- PODXL
-
- podocalyxin-like 1
-
- qRT-PCR
-
- quantitative real-time PCR
-
- SIX1
-
- Sine oculis homeobox homolog 1
-
- TMA
-
- tissue microarray
1 INTRODUCTION
Colorectal cancer is the third most common tumor and the third leading cause of cancer-related death in man and women in the world.1, 2 In China, there was an increase of over 376 300 cases of CRC and a total of 191 000 deaths during 2015.3 Environmental and lifestyle changes, including an altered diet, lack of appropriate physical activity, circadian disruption, and increase in alcohol consumption, have enormously aggravated the burden of CRC.4 In the past several years, the incidence and mortality of CRC increased rapidly and the onset age was much younger.5, 6 It is promising that therapeutic options for patients have increased substantially, including earlier diagnosis and treatments such as radiotherapy, chemotherapy, and surgical resection, however, the long-term survival of CRC patients remains unsatisfactory.7-10 Therefore, a deeper understanding of the underlying molecular mechanisms of CRC is urgently needed to accelerate the development of early detection strategies and effective therapeutic targets.
GINS complex subunit 4, also named SLD5, is a member of the tetrameric complex termed GINS, which is composed of PSF1, PSF2, PSF3, and GINS4. It is evolutionarily well-conserved and plays an essential role in the DNA replication initiation in eukaryotes.11, 12 GINS complex subunit 4 was also reported to be involved in early embryogenesis in mice and to maintain cell cycle progression and genome integrity in Drosophila.12-14 Prior studies suggested that GINS4 is highly expressed in lung cancer and facilitated lung cancer progression.15 Yamane et al16 reported that GINS4 expression was elevated both in bladder cancer tissues and cell lines, and the knockdown of GINS4 inhibited bladder cell growth both in vitro and in vivo. All of these findings suggested that GINS4 might play a significant role in tumorigenesis; however, the relevance of GINS4 in CRC has not been elucidated.
A large amount of evidence indicated that several transcription factors could inhibit cancer cell growth or migration.17 The zinc finger transcription factor KLF4 was reported to play an important role in tumor development and progression, and was considered as a potential tumor suppressor in some tumors; it can transcriptionally activate NDRG2 by binding with NDRG2 promoter and inhibit CRC cell growth through upregulating p21WAF1/Cip1 and downregulating cyclin D1.18 In addition, decreased KLF4 expression and increased Sp1 expression was a novel molecular mechanism of FOXM1c overexpression and that FOXM1c promoted cancer invasion and metastasis in human pancreatic cancer.19 Krüppel-like factor 4 played a negative role in gastric cancer cell invasion, which was reversed by upregulation of serine/threonine kinase 33 expression at the transcriptional level.20 Our previous studies found that loss of KLF4 expression contributed to enhance human gastric cancer EMT and metastasis development and progression through regulating PODXL.21 However, the underlying molecular mechanism of the tumor-suppressive role of KLF4 in CRC is still vague, and needs to be further investigated.
In the present study, GINS4 was highly expressed in CRC and downregulation of GINS4 could inhibit growth of CRC cells. Furthermore, KLF4 transcriptionally suppressed GINS4 expression in CRC, and the novel KLF4/GINS4 signaling pathway critically regulated CRC proliferation and growth to supply a promising prognostic indicator and an effective therapeutic target for CRC.
2 MATERIALS AND METHODS
2.1 Patients and specimens
Sixty-three paired fresh CRC and adjacent normal tissues were collected after radical surgical resection in Shanghai General Hospital from 2015 to 2017 and were stored at −80°C for RNA extraction. Additionally, 106 paired CRC and adjacent normal tissues were collected from patients diagnosed with CRC at the General Surgery Department of Shanghai General Hospital from 2013 to 2014. All specimens, to construct the TMA, were paraffin-embedded, validated by H&E staining, and finally examined by two independent pathologists. The final IHC results in TMA covered 106 CRC tissues and 108 adjacent normal tissues. None of the patients had received radiotherapy or chemotherapy before surgery. Clinicopathological characteristics were diagnosed and confirmed by two independent pathologists according to the guidelines of the American Joint Committee on Cancer, and are presented in Table 1. Written informed consent was obtained from each statement before enrolling in the study. The study was approved by the Ethical Committee for Clinical Research of Shanghai General Hospital.
| Parameter | Category | No. | GINS4 expression | χ2 | P value | ||
|---|---|---|---|---|---|---|---|
| Negative | Weak positive | Strong positive | |||||
| Age, years | <65 | 51 | 13 | 12 | 26 | 5.894 | .053 |
| ≥65 | 55 | 6 | 23 | 26 | |||
| Gender | Male | 55 | 10 | 21 | 24 | 1.612 | .447 |
| Female | 51 | 9 | 14 | 28 | |||
| T stage | T1 + T2 | 12 | 6 | 2 | 4 | 7.637 | .022* |
| T3 + T4 | 94 | 13 | 33 | 48 | |||
| N stage | N0 + N1 | 82 | 16 | 26 | 40 | 0.724 | .690 |
| N2 + N3 | 24 | 3 | 9 | 12 | |||
| TNM stage | I+II | 51 | 16 | 13 | 22 | 13.191 | .001 ** |
| III+IV | 55 | 3 | 22 | 30 | |||
| Nerve invasion | Yes | 27 | 5 | 8 | 14 | 0.191 | .909 |
| No | 79 | 14 | 27 | 38 | |||
| Vessel invasion | Yes | 40 | 6 | 17 | 17 | 2.618 | .270 |
| No | 66 | 13 | 18 | 35 | |||
| Differentiation | Well | 19 | 7 | 5 | 7 | 7.424 | .024 * |
| Moderate + poor | 87 | 12 | 30 | 45 | |||
| Tumor size, cm | <5 | 59 | 9 | 20 | 30 | 0.647 | .732 |
| ≥5 | 47 | 10 | 15 | 22 | |||
| Tumor location | Right | 41 | 9 | 13 | 19 | 0.740 | .691 |
| Left and rectal cancer | 65 | 10 | 22 | 33 | |||
| Tumor and normal | Tumor | 106 | 19 | 35 | 52 | 30.238 | .000 *** |
| Normal | 106 | 56 | 41 | 9 | |||
- Bold values indicate significance.
- * P < .05
- ** P < .01
- *** P < .001.
2.2 Cell lines and culture conditions
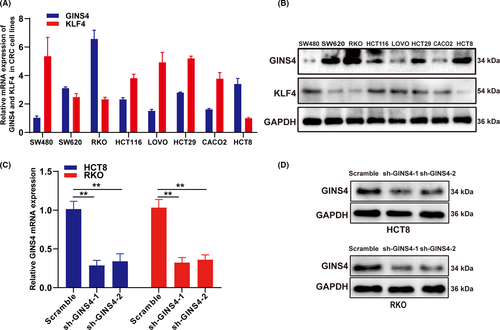
Human CRC cell lines (SW620, SW480, HCT29, RKO, CACO2, HCT116, LoVo, and HCT8) were purchased from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China). All cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) and maintained at 37°C in a humidified incubator with 5% CO2.
2.3 RNA extraction and qRT-PCR
Total RNA from CRC tissues and cells was extracted using RNAiso Plus Reagent (Takara) according to the manufacturer’s protocol. RNA (500 ng) was reverse transcribed with the PrimeScript RT reagent kit (Takara). cDNA was amplified with SYBR Premix Ex Taq (Takara). The following primers were used for qRT-PCR: GINS4, forward 5′-TCAAGCCTGTAATCCCAGCA-3′ and reverse 5′-GTTCAAGCGATTCTCCTGCC-3′; KLF4, forward 5′-AAAAAAGAATTCATGAGGCAGCCACCTG-3′ and reverse 5′-AAAAAAGTCGACTTAAAAATGCCTCTTCATGTG-3′; and GAPDH, forward 5′ GGGAAGGTGAAGGTCGGAGT 3′ and reverse 5′-GGGGTCATTGATGGCAACA-3′. The gene expression levels were determined by the 2–ΔΔCt method with GAPDH as the internal control mRNA. Each qRT-PCR was carried out in triplicate.
2.4 Protein extraction and western blot analysis
Total protein was lysed using RIPA buffer with the protease inhibitor PMSF (Beyotime Biotechnology), and the concentration of protein was measured with a BCA Protein Assay Kit (Tiangen). Protein lysates were separated by SDS-PAGE and then transferred onto PVDF membranes (Millipore). The membranes were blocked in 5% skim milk at room temperature for 1.5 hours followed by incubating with specific primary Abs at 4°C overnight: GINS4 (1:1000; Sigma), KLF4 (1:100; Santa Cruz Biotechnology), BCL2 (1:1000; Cell Signaling Technology), Ki67 (1:5000; Abcam), Cyclin D1 (1:10 000; Abcam), Erk1/2 (1:1000; Cell Signaling Technology), phospho-Erk (1:1000; Cell Signaling Technology), GAPDH (1:1000; Cell Signaling Technology), AKT (1:1000; Abcam), and phospho-AKT (1:1000; Abcam, USA). The membranes were then incubated with a HRP-conjugated anti-mouse or anti-rabbit secondary Ab (Cell Signaling Technology) at room temperature for 1.5 hours. After the membranes washed 3 times with TBST, protein bands were detected using ECL chemiluminescent reagent (Millipore).
2.5 Transient transfection and lentiviral transduction
For transient transfection, the pcDNA3.1-KLF4 plasmid and negative control vector were synthesized from Obio Technology. HCT8 and RKO cells cultured to 60%-70% confluence were transfected with the above oligonucleotides and vectors using Lipofectamine 2000 in accordance with the manufacturer’s instructions (Invitrogen). For lentivirus transfections, the shRNA plasmid for silencing GINS4 and the control-shRNA plasmid and were purchased from Genechem. The target sequences of the shRNA were as follows: sh-GINS4-1:5′-CCTATTTCCCATGATTCCTTCATA-3′; sh-GINS4-2:5′-GAGCAGAGGGACTACGTGATT-3′. HCT8 and RKO cells in logarithmic growth phase, cultured in 6-well plates, were transfected with lenti-shGINS4-virus or lenti-control virus. Then HCT8 and RKO cells with lentiviral transduction were purified with puromycin (Invitrogen). Cells transduced with control-shRNA plasmid (HCT8-scramble or RKO-scramble) served as controls. Gene silencing and overexpression were monitored by qRT-PCR and western blot analysis.
2.6 Cell growth assay
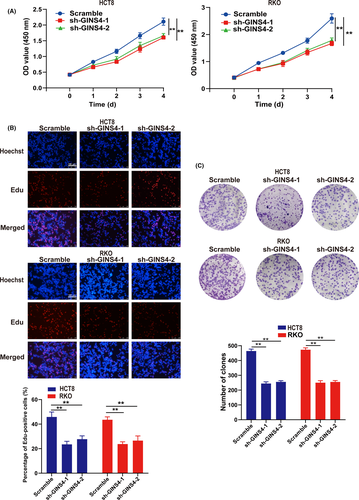
Cell plate colony formation assays were carried out to evaluate cellular growth in vitro. Cells were seeded into 6-well plates with 1000 cells/well and cultured at 37°C in a humidified atmosphere with 5% CO2. They were fixed in methyl alcohol for 15 minutes, then dyed with 0.1% crystal violet solution for 15 minutes after 14 days of culture. For the CCK-8 assay, 5000 cells/well were plated in 96-well cell culture plates. After 24 hours, 10 μL/well of CCK-8 (Dojindo) was added. The absorbance was measured at 450 nm on a Gen5 microplate reader (Bio-Tek). The CCK-8 assay was carried out at the indicated time point of every day for 4 days. For the EdU (RiboBio) cell growth assay, cells were plated into 24-well plates and cultured for 24 hours. The cells were incubated with 50 μmol/L EdU for 2 hours before fixation, permeabilization, and EdU staining. Cell nuclei were stained with Hoechst 33342 for 30 minutes. Images were captured using a fluorescence microscope (Leica). The percentage of EdU-positive cells was calculated as follows: (EdU-positive cells/Hoechst-positive cells) × 100%. All experiments were carried out in triplicate.
2.7 Flow cytometric analysis
For the cell cycle assay, CRC cells were harvested and fixed in 70% ethanol overnight at 4°C. After being washed with PBS, the cells were stained with 0.5 mL propidium iodide/RNase staining buffer at room temperature for 30 minutes. The cell apoptosis assay was undertaken using the Annexin V-PE/7-AAD Apoptosis Detection Kit (MultiSciences Biotech) according to the manufacturer’s protocol. Briefly, cells were suspended in 100 μL binding buffer, then 5 μL fluorochrome-conjugated annexin V-PE was added. After incubation for 15 minutes at room temperature, the cells were resuspended in 200 μL binding buffer and 10 μL 7-AAD was added. The apoptotic cells were analyzed by a flow cytometer (BD Biosciences).
2.8 Immunohistochemistry
The TMA was first deparaffinized in xylene, rehydrated in the gradient ethanol and prepared for antigen retrieval, then boiled in sodium citrate solution (0.01 M, pH 6.0) for 5 minutes to block endogenous peroxidase activity. The TMA was exposed to 3% hydrogen peroxide for 10 minutes, after being washed 3 times with PBS, and the slides were covered with primary Abs GINS4 (1:200; Sigma) and KLF4 (1:200; Santa Cruz Biotechnology) at 4°C overnight. The slides were incubated with the corresponding secondary Abs at 37°C for 0.5 hour and visualized using DAB to detect the primary Ab after incubation for 1 hour at room temperature.
The GINS4 and KLF4 intensity staining were scored as follows: 0, negative; 1, mild staining; 2, moderate staining; and 3, intense staining. The cell positive staining area was scored as follows: 0, <10%; 1, 10%-25%; 2, 26-50%; 3, 51%-75%) and 4, 76%-100%). The sum of intensity and extension was designated as the staining score, divided as follows: 0-3, negative expression; 4-6, weak positive expression; and 8-12, strong positive expression. Two pathologists blinded to patient information assessed staining scores independently to avoid bias.
2.9 Animal experiments
To investigate primary tumor growth, approximately 5 × 106 cells from stable cell lines (HCT8-shGINS4-1 and HCT8-scramble, and RKO-shGINS4-1 and RKO-scramble) were s.c. injected into 4-week-old male BALB/c athymic nude mice. Tumor volume was calculated once every 7 days using the following formula: volume = width2 × length × 0.5. All the mice were killed 5 weeks later, and xenografts were removed and weighed. All mice were used in line with the guidelines of Institutional Animal Care of Shanghai General Hospital.
2.10 Promoter reporter and dual-luciferase assay
Colorectal cancer cells were transfected with the full-length reporter plasmid (pGL4.27-GINS4-wild) or the mutated promoter (pGL4.27-GINS4-mut) with pCDNA3.1-KLF4 and pRL. The GINS4 promoter activity in these cells was normalized through cotransfection of a Renilla luciferase reporter containing a full-length Renilla luciferase gene. The resulting luciferase activity in the cells was quantified using a dual luciferase assay system (Promega) 36 hours after transfection. The effects of KLF4 on luciferase reporter plasmids were calculated with the ratio of firefly luciferase/Renilla luciferase activity. All experiments were independently repeated in triplicate.
2.11 Chromatin immunoprecipitation assay
The ChIP assay kit (Millipore) was prepared for ChIP assays in HCT8 and RKO cells (4 × 106) according to the manufacturer’s protocol, transfected with KLF4 or vector, after cross-linking, quenched with glycine, and protein and DNA were lysed by sonication. The supernatants were incubated overnight at 4°C with 2 μg anti-KLF4 (#AF3640; R&D Systems) and IgG served as a normal control. After purification, the resulting precipitated DNA fragments were analyzed using PCR to amplify fractions of the GINS4 promoter with the primers 5′-TCGTCCGATCCCAGGCTTCAAG-3′ (forward) and 5′- CCAGCGATCCTCCCACCTCAG-3′ (reverse). The DNA of PCR products were resolved electrophoretically on a 2% agarose gel and photographed using ultraviolet rays.
2.12 Statistical analysis
Data analyses were undertaken using SPSS 23.0 software. Paired and unpaired continuous variables were compared by Student’s t test or the Mann-Whitney U test. The correlations between GINS4 expression and clinicopathological features were determined by Fisher’s exact test and the χ2 test. The correlation between GINS4 and KLF4 expression was examined by Spearman’s correlation test. Survival curves were drawn using the Kaplan-Meier method and Cox multivariate analysis, and were analyzed by log-rank tests. In all of the tests, P < .05 was considered statistically significant.
3 RESULTS
3.1 Overexpression of GINS4 indicated poor clinical outcome of CRC
Using bioinformatics analysis, GINS4 was markedly increased both in CRC tissues and colon adenoma from the Oncomine database (https://www.oncomine.org) (Figure 1A). Thereafter, we sought to verify the expression of GINS4 in 63 paired CRC samples. Compared with the levels in adjacent normal mucosa tissues, the results of qRT-PCR indicated that the mRNA expression level of GINS4 was significantly upregulated in 49 (77.78%) fresh CRC clinical samples (Figure 1B). We undertook further IHC analyses with TMAs containing 106 cases of primary CRC and adjacent normal tissues. The clinicopathologic characteristics of the TMAs are listed in Table 1. We observed that GINS4 was positively stained in 88 (52 strong positive and 36 weak positive) cases among 106 specimens (Figure 1C). Western blot analysis further confirmed that GINS4 protein was elevated in CRC tissues (Figure 1D). Expression of GINS4 was positively associated with tumor stage (P = .022), differentiation (P = .024), and TNM stage (P = .001) (Figure1E-G, Table 1). However, there was no significant correlation between the level of GINS4 expression and age, gender, node stage, nerve invasion, vessel invasion, tumor size, or location (Table 1). The IHC results in TMA indicated that the level of GINS4 expression serves as a vital role in CRC development and progression. Our findings strongly illustrated that GINS4 plays a significant role in CRC development and progression.
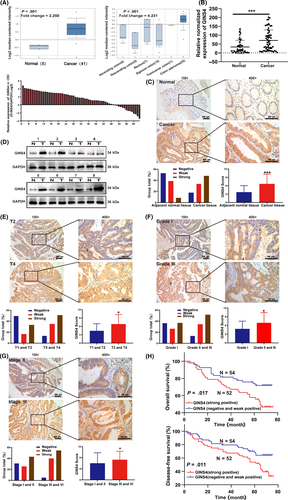
We then determined whether GINS4 acted as a valuable biomarker for the survival and prognosis of CRC patients. First, the univariate analysis indicated that patients with higher levels of GINS4 had poorer OS (P = .017) and DFS (P = .011) compared with those with lower levels of GINS4 (Figure 1H). The TNM stages and tumor location were significantly related to OS and DFS, based on the univariate analysis (Table 2). Second, according to the multivariate survival analysis based on the above factors in the Cox proportional hazards model, GINS4 expression was significantly related with OS (HR = 0.470; 95% CI, 0.248-0.892; P = .021) and DFS (HR = 0.486; 95% CI, 0.274-0.861; P = .013); TNM stage and tumor location were both closely related to OS and DFS (Table 2). These findings indicated that GINS4 played a vital role in CRC development and progression and could function as a valuable target for CRC therapy and diagnosis. Taken together, our findings suggested that GINS4 could serve as a valuable and unique indicator of CRC patient prognosis.
| Parameters | No. | OS | DFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Unvariate analysis | Multivariate analysis | Unvariate analysis | Multivariate analysis | ||||||
| χ 2 | P | HR(95%CI) | P | χ 2 | P | HR(95%CI) | P | ||
| Age | |||||||||
| <65 | 51 | 3.633 | .057 | 3.707 | .054 | ||||
| ≥65 | 55 | ||||||||
| Gender | |||||||||
| Male | 55 | 0.068 | .794 | 0.283 | .595 | ||||
| Female | 51 | ||||||||
| T stage | |||||||||
| T1+T2 | 12 | 1.132 | .287 | 2.286 | .131 | ||||
| T3+T4 | 94 | ||||||||
| N stage | |||||||||
| N0+N1 | 82 | 3.082 | .079 | 3.585 | .058 | ||||
| N2+N3 | 24 | ||||||||
| TNM stage | |||||||||
| I + II | 51 | 5.449 | .020* | 0.520 (0.275-0.983) | .044* | 5.459 | .019* | 0.556 (0.316-0.979) | .042* |
| III+IV | 55 | ||||||||
| Nerve invasion | |||||||||
| Yes | 27 | 2.478 | .115 | 2.986 | .084 | ||||
| No | 79 | ||||||||
| Vessel invasion | |||||||||
| Yes | 25 | 0.315 | .574 | 0.706 | .401 | ||||
| No | 81 | ||||||||
| Differentiation | |||||||||
| Well | 19 | 1.615 | .204 | 2.375 | .123 | ||||
| Moderate+Poor | 87 | ||||||||
| Tumor size | |||||||||
| <5cm | 59 | 0.076 | .783 | 0.029 | .866 | ||||
| ≥5cm | 47 | ||||||||
| Tumor location | |||||||||
| Right | 41 | 4.927 | .026* | 2.125 (1.152-3.921) | .016* | 5.366 | .021* | 2.072 (1.192-3.600) | .010* |
| Left and Rectal cancer | 65 | ||||||||
| GINS4 expression | |||||||||
| Negative and weak positive | 54 | 5.733 | .017* | 0.470 (0.248-0.892) | .021* | 6.446 | .011* | 0.486 (0.274-0.861) | .013* |
| Strong positive | 52 | ||||||||
- Abbreviations: HR: hazard ratio; CI: confidence interval.
- * P < .05.
3.2 Knockdown of GINS4 expression inhibits growth in vitro and in vivo
To investigate the function of altered GINS4 expression on the growth of CRC cells, GINS4 was notably upregulated in RKO and HCT8 cell lines compared with other CRC cell lines, through qRT-PCR and western blot analysis (Figure 2A,B). Hence, we chose HCT8 and RKO cells to establish stable cell lines to explore the influence of GINS4 expression in the biological processes of CRC cells. HCT8 and RKO cells transfected with lentiviral shGINS4-1 or shGINS4-2 vector were applied in this study, and western blotting and real-time PCR were used to evaluate the efficiency of shGINS4-1 and -2. The results showed that shGINS4-1 and -2 effectively blocked GINS4 protein (Figure 2C,D). The CCK-8 and EdU growth assay results indicated that low levels of GINS4 expression inhibited RKO and HCT8 growth (Figure 3A,B). Furthermore, the colony formation assay showed that GINS4 knockdown inhibited colony formation both in HCT8 and RKO cells (Figure 3C). In the s.c. tumor model, the 4 stable cell lines HCT8-shGINS4-1/HCT8-scramble and RKO-shGINS4-1/RKO-scramble were s.c. injected into nude mice. After 10 days, the tumor volume was monitored twice every week. The results showed that the volume and weight of the xenograft tumor from HCT8-shGINS4-1/RKO-shGINS4-1 cells were remarkably smaller than those in the HCT8-scramble/RKO-scramble group (Figure 4A-C). Furthermore, IHC analysis showed that GINS4 expression was significantly downregulated in HCT8-shGINS4-1/RKO-shGINS4-1 tumors (Figure 4D). Our findings primarily indicated that GINS4 served as an oncogene and knockdown of GINS4 expression can inhibit the growth of CRC cells in vitro and in vivo.

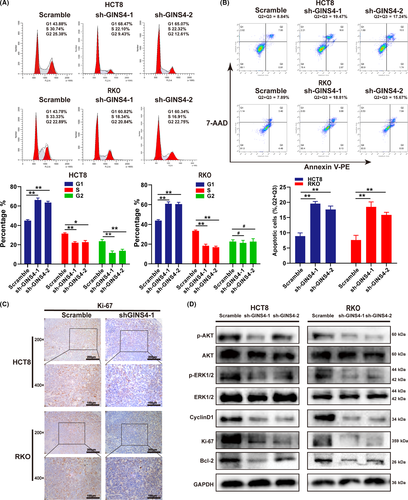
3.3 Knockdown of GINS4 suppresses cell cycle progression and promotes cell apoptosis by inhibiting PI3K/AKT and MAPK/ERK pathways
As we know, equilibrium between cell proliferation and cell apoptosis is important for cell growth. To explore how GINS4 regulates CRC cell growth, we investigated whether the lower expression of GINS4 inhibited the transition from G0/G1 phase to S phase (Figure 5A). Downregulated expression of GINS4 remarkably reduced the expression of G1/S phase transition regulator Cyclin D1 and Ki-67 in HCT8 and RKO cells (Figure 5D)22. In the s.c. tumor model, IHC and western blot analyses showed that cell proliferation marker Ki-67 was significantly downregulated in HCT8-shGINS4-1/RKO-shGINS4-1 tumors (Figures 4B and 5C). As expected, determined by annexin V-PE/7-AAD apoptosis assay, GINS4 also inhibited CRC cells apoptosis. Furthermore, the antiapoptotic protein Bcl-2 was decreased in HCT8 and RKO cells when expression of GINS4 was downregulated (Figure 5B,D). Our previous study indicated that GINS4 inhibited cell apoptosis in vitro and promoted cell growth and metastasis in vitro and in vivo in gastric cancer; GINS4 directly combined and activated Rac1/CDC42, thereby activating their downstream pathways, including the PI3K/AKT, MAPK/ERK, and PTEN pathways in gastric cancer.23 We examined the activity of the 3 signaling pathway cascades, and found that GINS4 knockdown only decreased the expression of pERK and pAKT in CRC cell lines (Figure 5D). Therefore, the activity of MAPK/ERK and PI3K/AKT pathways was suppressed when GINS4 was downregulated (Figure 5D). These results indicated that GINS4 knockdown affected cell proliferation and apoptosis through inhibiting the MAPK/ERK and PI3K/AKT pathways in CRC cells.
3.4 Krüppel-like factor 4 downregulates GINS4 expression by directly binding to its promoter
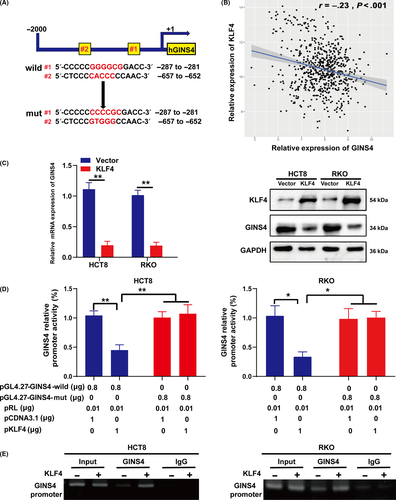
To explore the mechanisms underlying upregulated GINS4 in CRC tissues and cell lines, the sequences of GINS4 gene promoter for potential transcription factor binding sites were analyzed by using the JASPAR (http://jaspar.genereg.net/) database and Alggen-PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). The results showed that there were 2 putative KLF4-binding sites in the GINS4 promoter region containing potential KLF4-binding elements, 5′-CACCC-3′ and 5′-(G/A)(G/A)GG(C/T)G(C/T)-3′ (Figure 6A).24, 25 Furthermore, statistical analysis of The Cancer Genome Atlas datasets revealed that the expression of GINS4 was negatively correlated with the expression of KLF4 in the 663 CRC specimens (Figure 6B); a negative correlation between GINS4 and KLF4 in CRC cell lines was already determined (Figure 2A,B). We elevated the expression of KLF4 in CRC cells, and the results indicated that the overexpression of KLF4 in HCT8 and RKO cells led to decreased expression of GINS4 mRNA and protein (Figure 6C). To further confirm whether KLF4 could transcriptionally inhibit GINS4, the full-length promoter of GINS4 was cloned to pGL4.27 vector, and we also mutated the 2 binding sites (Figure 6A). Furthermore, we cotransfected pGL4.27-GINS4-wild or pGL4.27-GINS4-mutant with pCDNA3.1-KLF4 or control into HCT8 and RKO cells. We found that KLF4 prominently suppressed the wild GINS4 promoter activity but not mutant GINS4 promoter activity in HCT8 and RKO cells (Figure 6D). To explore whether KLF4 directly regulates GINS4 expression, we used the ChIP assay and observed that KLF4 indeed bound to the GINS4 promoter in HCT8 and RKO cells (Figure 6E). In summary, these results showed that KLF4 downregulated GINS4 expression by directly binding to GINS4 promoter.
3.5 Combination of KLF4 and GINS4 levels shows prognostic ability in CRC patients
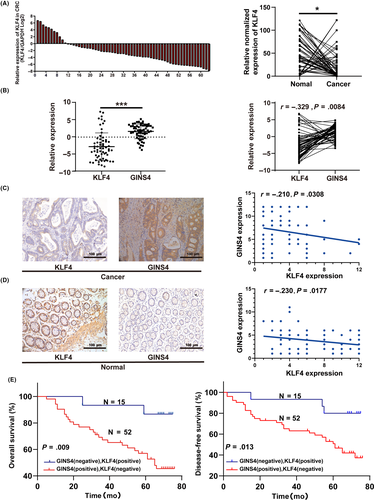
To further verify the correlation between the expression of KLF4 and GINS4 in the CRC cases, we undertook qRT-PCR to analyze KLF4 expression in the same CRC specimens used in GINS4. The results showed that the level of KLF4 was lower in CRC tissues compared to normal colorectal mucosa tissues (Figure 7A). Further analysis showed that the correlation between GINS4 and KLF4 in CRC tissues compared to normal colorectal mucosa tissues was negative, with statistical significance (Figure 7B). Furthermore, we investigated the expression of KLF4 with the existing CRC TMA used to analyze GINS4 expression, described above. The results showed that the CRC specimens in the TMA, which lost expression of KLF4, had high expression of GINS4, whereas the opposite results were found in normal colorectal mucosa tissues (Figure 7C,D). Statistical analysis revealed that expression of KLF4 was weakly negatively correlated with GINS4 both in normal colorectal mucosa and CRC tissues (Figure 7C,D). Moreover, pairwise comparisons indicated that patients with high KLF4 (positive) and low GINS4 (negative) staining levels had significantly better OS and DFS than patients with low KLF4 (negative) and high GINS4 (positive) staining (Figure 7E). This indicated that the combination of KLF4 and GINS4 levels could be a useful predictor of clinical prognosis in CRC patients.
4 DISCUSSION
In this report, it was found that GINS4 served as a significant prognostic biomarker and promoted CRC growth by inhibiting cell apoptosis and accelerating cell cycle and colony formation progression, which are the vital characteristics of cancer progression. As a tumor suppressor in CRC was reported in many previous research,18, 26 KLF4 downregulated GINS4 at the transcriptional level by directly binding to the GINS4 promoter domain region, establishing a novel KLF4/GINS4 signaling pathway that regulated growth of CRC. Taken together, we showed that lost expression of KLF4, a transcriptional repressor of GINS4, resulted in overexpression of GINS4 and ultimately promoted growth and proliferation of CRC.
GINS complex subunit 4 is a member of the GINS family of proteins that plays a vital role in initiation of DNA replication and elongation, and also accelerates the G1/S phase transition in the cell cycle.27, 28 Elevated expression of PSF1 (a member of the GINS family) has been confirmed in different cancers, eg, in cancer-initiating cells or cancer stem cells related to a mice tumor cell transplantation model with high PSF1 promoter activity.16, 29 In this study, we proved that GINS4 was an oncogene from different aspects. First, GINS4 expression was markedly increased both in CRC tissues and cell lines. Second, further analysis of the correlation between TMA clinicopathologic characteristics of the CRC patients from whom the TMA specimens were achieved indicated that GINS4 expression was positively related with CRC advanced tumor stage, poor differentiation, and TNM stage. Univariate and multivariate survival analyses indicated that CRC patients with higher GINS4 expression experienced low OS and DFS. Third, we generated stable CRC cell lines to undertake in vitro and in vivo assays. Using CCK-8, EdU, and colony formation assays, we found that the GINS4 knockdown inhibited the growth of CRC cells, which was also verified by in vivo tumor xenograft assays, and that GINS4 knockdown inhibited growth and the cell cycle and accelerated cell apoptosis progression in vitro, using flow cytometric analyses. Finally, our results were consistent with a previous study in which elevated expression of GINS4 was detected in human bladder cancer cells, and GINS4 knockdown substantially suppressed bladder cancer cell growth.16 In summary, our findings underscored that GINS4 serves as a tumor oncogene that could promote CRC growth.
The canonical ERK1/2 signal pathway, known as a member of the MAPK family, is often deregulated in many cancers.30, 31 Activation of ERK1/2 was reported to stimulate cell growth and inhibit cell apoptosis through many mechanisms, including the maintenance of activated Cyclin D1 and enhanced AP-1 activity.32, 33 The PI3K/AKT signaling pathway is also one of the most frequently activated pathways and modulates cell growth and apoptosis in various tumors.34, 35 A novel study reported that the PI3K/AKT signaling pathway increased C-myc and facilitated cell growth in CRC.36 Here, our results showed that GINS4 knockdown simultaneously trigged apoptosis and cell cycle arrest and suppressed the MAPK/ERK and PI3K/AKT signaling pathways, which are 2 main downstream pathways of GINS4 identified in our previous study.23
Although the molecular basis of GINS4 overexpression in cancer cells is still vague, a recent study showed that GINS4 protein could be stabilized by lymphoid-specific helicase in nonsmall-cell lung cancer.13 Transcription factors played a significant role in the development of tumors. Our recent study suggested that SIX1 could promote cancer cell migration and invasion through increasing vimentin expression at the transcriptional level directly by binding to the promoter domain of vimentin in gastric cancer.37 Our previous studies found that KLF4 was downregulated both in gastric cancer and pancreatic cancer and regulated the expression of a vast number of oncogenes that are involved in many cellular functions, ranging from differentiation to growth, metastasis, and EMT. Loss of KLF4 expression contributed to human gastric cancer EMT and metastasis by regulating PODXL.21 Krüppel-like factor 4 suppressed pancreatic cancer EMT and metastasis by reducing the expression of caveolin-1 through directly binding the promoter region of caveolin-1.38 Furthermore, KLF4 expression was at a significantly low level and loss of KLF4 was an independent predictor of survival and recurrence in colon cancer,39 and an interferon-inducible gene, IFITM3, could be negatively regulated by KLF4 at the transcription level in colon cancer.40 In the present study, qRT-PCR, western blot, the IHC results in TMA combined with our clinical data indicated that KLF4 was downregulated in CRC tissues compared with paired adjacent normal tissues. We found that overexpression of KLF4 caused low levels of GINS4 in CRC. Furthermore, bioinformatics analysis from JASPAR and Alggen-PROMO predicted that there were 2 binding sites in the GINS4 promoter related to the putative KLF4 binding motif; we synthesized the wild full-length promoter of GINS4 and mutated the 2 binding sites. Luciferase reporter assay confirmed that KLF4 can bind to particular GINS4 promoter elements and transcriptionally inhibit GINS4 expression. Downregulation of miR-370 could suppress GINS4 expression in bladder cancers,16 and our recent study showed that circMLLT10 served as a miR-509-3-5p sponge and promoted gastric cancer cell growth and metastasis by increasing GINS4 expression and activating Rac1 and CDC42.23 Therefore, the relationships between noncoding RNA and KLF4 in regulating GINS4 require further study, which might contribute to exploring the progression of CRC.
In summary, based on our clinical and experimental evidence, we highlighted that GINS4 was a vital oncogene in CRC and found that its expression was remarkably elevated in both CRC cell lines and clinical specimens. Elevated GINS4 expression promoted CRC growth in vitro and in vivo and it was transcriptionally inhibited by KLF4, which served as a crucial tumor-suppressive transcriptional factor in CRC. Fundamentally, we not only determined that GINS4 might act as a valuable prognostic biomarker of CRC, but also revealed the aberrant KLF4/GINS4 signaling pathway in CRC, which could provide significant new insights into the development of novel therapies for CRC.
ACKNOWLEDGMENTS
This work was ratified by the Ethics Committee of Shanghai General Hospital. All animals were used according to the Shanghai General Hospital Animal Care and Use Guidelines. Written informed consents was obtained from all patients. This work was supported by grants from the National Natural Science Foundation of China (817725276), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20161425), Shanghai Jiaotong University Medical Cross Fund (YG2017MS28), Shanghai Songjiang Science and Technology Project Fund (18SJKJGG23 and 19SJKJGG22).
DISCLOSURE
The authors declare no conflicts of interest in this article.




