p62 promotes bladder cancer cell growth by activating KEAP1/NRF2-dependent antioxidative response
Abstract
p62 is associated with 2 major cellular defense mechanisms against metabolic and oxidative stress, autophagy and the Kelch-like ECH-associated protein 1 (KEAP1)-nuclear factor-E2-related factor 2 (NRF2) system. Recent studies indicate that the p62-KEAP1-NRF2 pathway promotes tumorigenesis and tumor growth mediated by NRF2-dependent antioxidative response. However, whether p62 is involved in bladder cancer (BCa) development remains unknown. Here, we found that p62 is overexpressed in BCa tissue and several BCa cell lines. The knockdown of p62 inhibits BCa cell growth both in vitro and in vivo, with increased intracellular reactive oxygen species level. Mechanically, p62 activates NRF2 signaling by sequestrating KEAP1, which leads to the upregulation of antioxidant genes (Gclc, Gstm5, and Gpx2), thus protecting BCa cells from oxidative stress. Our findings indicate that p62 might be involved in the development of BCa and serve as a potential therapeutic target.
1 INTRODUCTION
Bladder cancer (BCa) is the most common urinary tract cancer, accounting for 549 000 new cases and 200 000 deaths worldwide in 2018.1 In the past decade, the incidence and mortality of BCa in China have gradually increased.2 Currently, the most common treatments for BCa are surgery, radiotherapy, and chemotherapy. Due to the characteristics of easy recurrence, invasion, and metastasis, there has been no technical advancement or innovation in the treatment of BCa. Therefore, an in-depth study of the molecular mechanisms of BCa and search for new drug therapeutic targets have become urgently needed.
p62, encoded by Sqstm1, is a multifunctional stress-inducible scaffold protein involved in diverse cellular processes.3 p62 is associated with 2 major cellular defense mechanisms against metabolic and oxidative stress, autophagy and the Kelch-like ECH-associated protein 1 (KEAP1)-nuclear factor (NF)-E2-related factor 2 (NRF2) system. p62 serves as a selective autophagy adaptor and plays a key role in regulating the formation of protein aggregates. In addition, p62 activates the KEAP1-NRF2 system by sequestering KEAP1 through its KEAP1-interacting region (KIR) domain.4, 5 p62 is also linked to other cellular processes through its interaction with many intracellular signalings, including NF-κB6 and mTOR.7 Mounting evidence has indicated that p62 is associated with a wide range of diseases, including various cancers, such as liver,8 lung,9 breast,10 pancreatic,11 prostate,12 and colon.13 For example, p62 promotes hepatocellular carcinogenesis by protecting liver cancer-initiating cells from oxidative stress.14 However, the role of p62 in BCa remains unknown.
Nuclear factor-E2-related factor 2 is a master regulator of oxidative response. Under the basal condition, NRF2 is tightly controlled by KEAP1.15 Intracellular oxidation prevents KEAP1-mediated proteasomal degradation of NRF2. Then NRF2 translocates into the nucleus and induces antioxidant-related genes, which help to protect cells from oxidative stress. Intriguingly, Sqstm1 is a target gene of NRF2. Sqstm1 protein, namely p62, interacts with KEAP1 and inhibits KEAP1-mediated NRF2 degradation, leading to the stabilization and activation of NRF2.4 Accumulating evidence has established the NRF2 pathway as a driver of cancer progression, metastasis, and resistance to therapy.16
Here, we observed the overexpression of p62 in human BCa tissue and several BCa cell lines. We found that knockdown of p62 increased intracellular reactive oxygen species (ROS) levels and inhibited BCa cell growth. Mechanically, p62 activates the NRF2-dependent antioxidative pathway by sequestrating KEAP1, thus protecting BCa cells from oxidative stress. Our findings thus indicate that p62 might promote the development of BCa by regulating intracellular antioxidant response.
2 MATERIALS AND METHODS
2.1 Cell culture and transfection
T24, RT4, 5637, TCCSUP, 253J, SV-HUC-1, and SW780 cell lines were purchased from ATCC, and all BCa cells were cultured in DMEM or RPMI-1640 medium supplemented with 10% FBS. The single guide RNAs (sgRNAs) targeting p62 (sgp62) were constructed in this study. The upstream sequence of sgp62 was 5′-CACCGCATTGTCAATTCCTCGTCAC-3′, and the downstream sequence was: 5′-AAACGTGACGAGGAATTGACAATGC-3′ (sequence #1). The upstream sequence of sgp62 was 5′-CACCGAATGGCCATGTCCTACGTGA-3′, and the downstream sequence was: 5′-AAACTCACGTAGGACATGGCCATTC-3′ (sequence #2). Single guide RNA transfections were applied to silence the expression of p62 in BCa cells. p62 cDNA was cloned into the pcDNA3.1 vector. The sgRNAs or plasmids, Lipofectamine 2000 (Invitrogen) and FBS-free medium, were mixed according to the manufacturer’s protocol for transfection. 293FT cells were applied for the packaging of lentivirus. Real-time PCR and western blot analyses were used to monitor transfection efficiency.
2.2 Reagents and Abs
Cycloheximide (CHX) and bafilomycin A1 (Baf A1) were purchased from Sigma-Aldrich. Rabbit primary Abs against p62, B-cell lymphoma 2 (Bcl-2)-associated X protein (Bax), and KEAP1 were purchased from Cell Signaling Technology. Mouse primary Abs against Bcl-2 and β-actin were purchased from Cell Signaling Technology. The rabbit primary Abs against NRF2 and glutamate-cysteine ligase catalytic subunit (GCLC) were purchased from Abcam.
2.3 Colony formation assay
After sgRNA infection and selection, the cells were plated in 6-well plates at a density of 1000 cells/well. After 10 days of culture, the cells were washed with PBS, fixed with 4% paraformaldehyde, and stained with crystal purple for 10 minutes. The stained colonies were then photographed and counted.
2.4 Cell proliferation assay
The BCa cells were inoculated into 96-well plates at a cell density of 1 × 104 cells per well and incubated at 37℃, 5% CO2 atmosphere overnight. Each well was added with MTT at 0, 24, 48, and 72 hours. After incubating for another 4 hours, DMSO was added to each well. The crystals were sufficiently dissolved by shaking for 10 minutes, and the absorbance was measured at a wavelength of 490 nm to calculate the cell survival rate.
2.5 Western blot analysis
Bladder cancer cell protein extracts were prepared according to standard protocols. Cell lysates were separated by 10% SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were incubated with specific primary Abs at 4℃ overnight. The protein immunoreactive signals were tested by the ECL detection system (Thermo Fisher Scientific).
2.6 Immunofluorescence
5637 Cells were plated on coverslips, fixed with 4% paraformaldehyde for 20 minutes, and permeabilized in PBS containing 0.3% Triton X-100 for 10 minutes. Cells were then incubated in the blocking buffer (5% BSA in PBS) at room temperature for 1 hour followed by staining with primary Abs (p62 and KEAP1). Coverslips were washed and incubated for 1 hour with the secondary Abs. Next, the cells were stained with DAPI and blocked with glycerol. Fluorescence microscopy was applied to detect the fluorescence of the cells.
2.7 Immunohistochemistry
Tissue specimens were fixed with 4% paraformaldehyde and embedded with paraffin for further analysis. The sections were stained with the specific primary Abs as described above. Images of tumor slides were obtained on a Nanozoomer Slide Scanner (Hamamatsu). Bladder cancer tissues were obtained from patients undergoing resection at the First Affiliated Hospital of Xi’an Jiaotong University. Written informed consent was obtained from the patients, and the protocol was approved by the Ethical Review Board of Xi’an Jiaotong University.
2.8 Quantitative RT-PCR
Total RNA was extracted using the RNeasy Kit (Qiagen) according to the manufacturer’s protocol. Then, 1 μg total RNA was reverse transcribed into cDNA using a reverse transcription kit (Takara). The cDNA was used for quantitative PCR analysis on an iCycler iQ5 Real-Time PCR detection system (Bio Rad) following the manufacturer’s instructions. The expression of the target gene was normalized to the expression of Gapdh. Relative gene expression was calculated using the standard 2−ΔΔCt method. The following primer sequences are used: Gclc F, 5′-GGGGTGACGAGGTGGAGTA-3′; R, 5′-GTTGGGGTTTGTCCTCTCCC-3′; Gstm5 F, 5′-TCATCCAAGTCTATGGTTCTGGG-3′; R, 5′-CCACAGATGTACCGTTTCTCCT-3′; Gpx2 F, 5′-GCCTCAAGTATGTCCGACATG-3′; R, 5′-GGAGAACGGGTCATCATAAGGG-3′; and Gapdh F, 5′-GCTAAGCAGTTGGTGGTGCA-3′; R, 5′-TCACCACCATGGAGAAGGC-3′.
2.9 Intracellular ROS and glutathione detection
Intracellular ROS were detected by 2′,7′-dichlorofluorescin diacetate (DCFDA; Applygen). Briefly, BCa cells were stained with 5 μmol/L DCFDA in an incubator at 37°C for 30 minutes then washed twice in PBS before FACS analysis. Intracellular glutathione (GSH) was measured with a colorimetric GSH detection kit (Solarbio). At least 106 cells were washed with ice-cold PBS and lysed in lysis buffer. After 2-3 freeze-thaw cycles in liquid nitrogen, the supernatant was collected for measurement of absorbance at 412 nm. The GSH standard curve was generated for determining the sample GSH concentrations.
2.10 Bladder cancer xenografts
5637 Cells were stably infected with sgp62 and control (sgCon). Male Balb/c nude mice at the age of 5-6 weeks were purchased from the Laboratory Animal Center of Xi'an Jiaotong University. The sgp62 5637 or sgCon 5637 cells (1 × 106) were injected s.c. into the right flank. Tumor volumes were calculated using the formula 0.5 × ab2 (“a” = length, “b” = width). All animal experiments were carried out according to the guidelines approved by the Animal Care and Use Committee of the Laboratory Animal Center of Xi’an Jiaotong University.
2.11 Statistical analysis
Statistical analysis was undertaken with GraphPad Prism version 5 software. Data are presented as mean ± SEM. Student’s t test was used for data normally distributed. A P value of less than .05 was considered statistically significant.
3 RESULTS
3.1 p62 is overexpressed in human BCa and cell lines
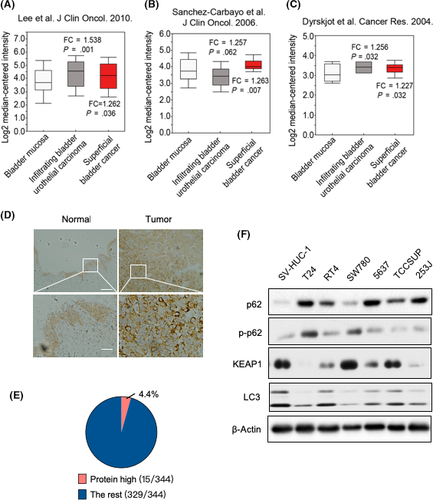
To explore the role of p62 in BCa, we first analyzed the p62 expression in human BCa. p62 was found to be overexpressed in 3 different BCa studies (Figure 1A-C).17-19 These 3 studies obtained gene expression data from 165,17 109,18 and 4119 BCa samples, respectively, by microarray analysis. Furthermore, analysis of clinical tissue specimens confirmed the upregulation of p62 in BCa tissues (Figure 1D). We also noticed that the protein level of p62 in BCa patients upregulated (more than twofold) at a frequency of 4.4% (15 of 344 cases) in The Cancer Genome Atlas database (Figure 1E).20 We then determined the expression of p62 in several human BCa cell lines by western blots. Among 7 cell lines tested, T24, RT4, 5637, TCCSUP, and 253J showed high p62 baseline expression, whereas p62 was barely detected in SV-HUC-1 and SW780 (Figure 1F). These data suggest that p62 was upregulated in human BCa, at least partially.
3.2 p62 promotes BCa cell proliferation and inhibits its apoptosis in vitro
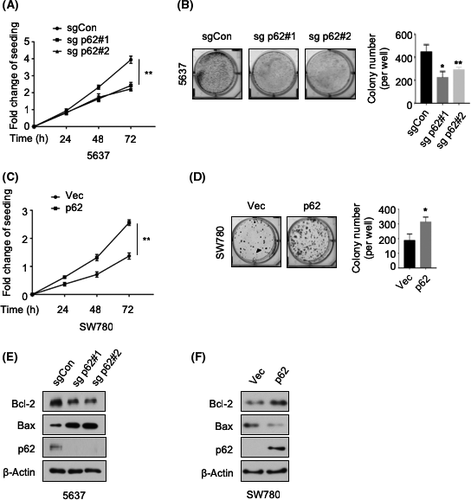
To study the biological function of p62 in BCa, we applied the CRISPR/Cas9-mediated gene knockdown strategy in 5637 cells, which showed a high p62 baseline expression (Figure 1F). Both MTT cell proliferation study and colony formation assay showed that p62 deletion strongly inhibited cell growth of 5637 (Figure 2A,B). In addition, we took advantage of the SW780 cell line, which expressed a low level of p62 (Figure 1F), to overexpress p62. We observed that p62 overexpression significantly promoted the growth of SW780 cells (Figure 2C,D). Furthermore, p62 knockdown decreased the level of Bcl-2 while it increased the level of Bax, indicating a proapoptotic phenotype of p62-deficient BCa cells (Figure 2E). In contrast, p62 overexpression induced the opposite effect (Figure 2F). Together, these data indicate a requirement for p62 in BCa cell growth.
3.3 p62 is required for oxidative resistance in BCa cells
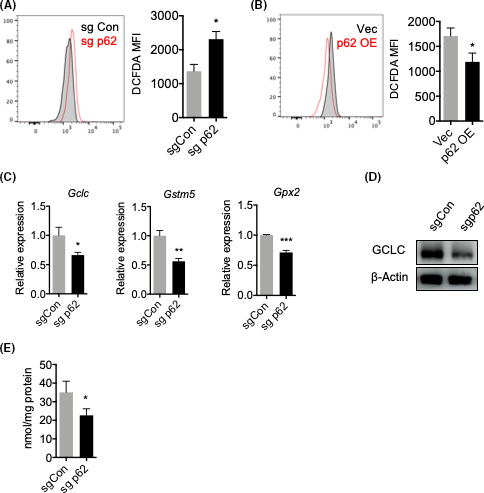
As p62-NRF2 signaling is the major intracellular antioxidative response,3 we speculated that p62 is related to the redox homeostasis within BCa cells. To that end, we detected intracellular ROS levels by a DCFDA probe. Indeed, we found that p62 deletion resulted in an increased level of ROS (Figure 3A), whereas p62 overexpression rescued this phenotype (Figure 3B). We then investigated the activation of NRF2 by determining the expression of NRF2 targeted genes, among which we observed a significant downregulation of Gclc, Gstm5, and Gpx2 after p62 knockdown (Figure 3C,D). More importantly, p62 knockdown led to a decrease in GSH, one of the major antioxidants to prevent oxidative stress (Figure 3E). These results suggest that p62 activates the NRF2-dependent antioxidative response in BCa cells, thus protecting them from oxidative stress and promoting their growth.
3.4 p62 activates NRF2 by sequestering KEAP1
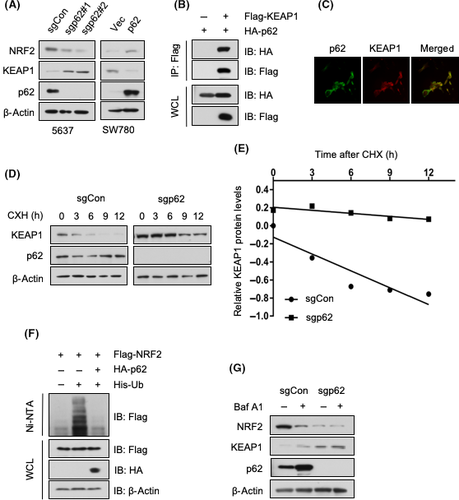
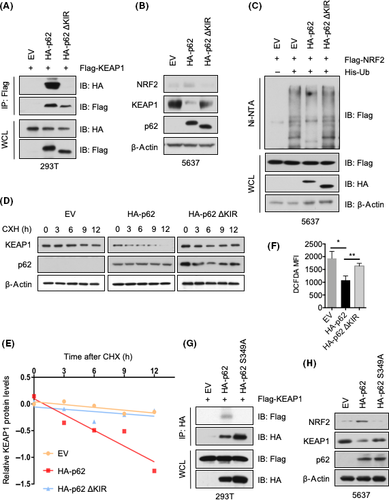
We then attempted to obtain the molecular mechanism by which p62 activates NRF2. As mounting evidence indicates that p62 is a crucial regulator of the KEAP1/NRF2-dependent antioxidative response,3 we asked whether p62 participates in this pathway in BCa cells. p62 deletion in 5637 cells resulted in an elevated level of KEAP1 and, at the same time, a decreased level of NRF2. Overexpression of p62 in SW780 cells led to the opposite effects (Figure 4A). This is consistent with previous studies that reported that KEAP1 is degraded through p62-mediated autophagy.21, 22 We then confirmed the interaction between p62 and KEAP1 by coimmunoprecipitation in 293T cells (Figure 4B). Moreover, p62 and KEAP1 were colocated in 5637 cells (Figure 4C). To evaluate the role of p62 in regulating KEAP1 protein abundance, we undertook the CHX chase assay in 5637 cells. We found that p62 deletion slowed down the KEAP1 degradation rate (Figure 4D,E). Furthermore, p62 inhibited the ubiquitylation of NRF2 (Figure 4F). As p62 is degraded through autophagy, we used Baf A1 to induce autophagy impairment, which led to the accumulation of p62 (Figure 4G). Consistent with p62 overexpression (Figure 4A), autophagy inhibition increased the level of KEAP1 while it decreased the level of NRF2 in 5637 cells (Figure 4G). These data indicate that p62 in BCa cells activates NRF2 by regulating KEAP1 abundance.
It has been reported that p62 activates NRF2 signaling by sequestering KEAP1 through its KIR domain.5 To determine whether the KIR domain mediates the interaction with KEAP1, we generated a KIR-deleted mutant of p62, then examined the interaction of the mutant with KEAP1 by assessing their immunoprecipitation. We observed that the KIR-deleted p62 failed to precipitate together with KEAP1 (Figure 5A). Moreover, the KIR-deleted mutant restored the KEAP1 level and decreased NRF2 abundance (Figure 5B) by increasing the ubiquitination of NRF2 (Figure 5C). In addition, the CHX chase assay showed a KIR-dependent degradation of KEAP1 (Figure 5D,E). Consistently, the KIR-deleted p62 failed to reduce ROS in BCa cells, indicating the deficiency of a potent intracellular antioxidant response (Figure 5F). It has been reported that the phosphorylation of p62 at serine 349 is important for the p62-mediated NRF2 activation.4 We confirmed this mechanism in the BCa cells by using a p62 S349A mutant, which failed to interact with KEAP1 (Figure 5G) and induce NRF2 activation (Figure 5H). In fact, we observed a basal level of phospho-p62 in all 7 cell lines (Figure 1F). These results collectively suggest that the p62 KIR domain and p62 phosphorylation at S349 are both important to p62-KEAP1 interaction, which disrupts the KEAP1-NRF2 association and provokes NRF2 stabilization and activation.
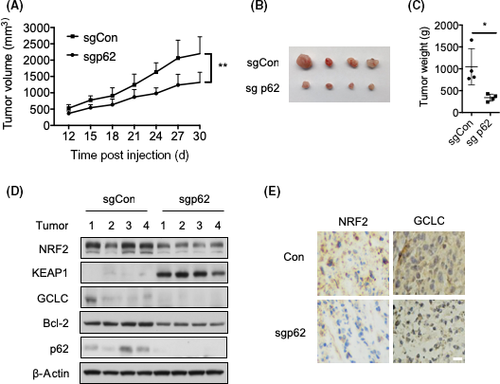
3.5 Knockdown of p62 inhibits BCa growth in vivo
Finally, we examined the role of p62 in BCa cell growth in vivo. p62-depleted 5637 cells were inoculated into nude mice. Tumor volume (Figure 6A,B) and weight (Figure 6C) were dramatically reduced when p62 was deleted. Moreover, the deficiency of p62 led to the upregulation of KEAP1 and downregulation of NRF2 (Figure 6D), thus blunting KEAP1-NRF2 signaling, which was consistent with the in vitro data (Figure 4A). Accordingly, one of the NRF2-targeted genes, GCLC, was downregulated (Figure 6D). In addition, the level of Bcl-2 was decreased in p62-depleted 5637 cells (Figure 6D). Consistently, immunohistochemistry results showed that NRF2 and GCLC were also reduced in p62-depleted 5637 cells (Figure 6E). Together, our data suggest that p62 promotes BCa cell proliferation by activating NRF2-dependent antioxidative response in vivo.
4 DISCUSSION
Although mounting evidence suggests that p62 is involved in tumor initiation and progression,3 little is known about its role in BCa. Our study showed that p62 is overexpressed in a portion of BCa patients and BCa cell lines. The in vivo and in vitro data indicated that p62 promotes BCa cell growth by activating KEAP1-NRF2 signaling and protecting cancer cells from oxidative stress.
It is well appreciated that p62 is involved in tumor development. Some of the first evidence is that accumulation of p62 during tumorigenesis is associated with the upregulation of NF-κB, a critical survival regulator induced by the oncogene Ras.23 In addition, p62 facilitates the tumor growth through the activation mTORC1 and induction of c-Myc.14 Dysfunction of the p62-KEAP1-NRF2 axis has been observed in several cancer types.3 For instance, persistent phosphorylation of p62 at serine 349 has been found in hepatoma,8, 24 which results in the activation of NRF2.4 Knockout of p62 in a liver cancer cell line markedly abrogates tumor growth.24 We found that upregulation of p62 promotes NRF2 activation in BCa cells, in which the phosphorylation of p62 at serine 349 might play a crucial role. In addition, p62 is a selective receptor of autophagy, which has been reported to be associated with genomic damage, metabolic stress, and tumorigenesis.25 Autophagy plays dual roles in cancer biology. The basal level of autophagy operates as a mechanism for tumor suppression through the reduction of damaged cellular components.26 However, autophagy also helps cancer cells to overcome stressful conditions (such as hypoxia and nutrient deprivation) by recycling intracellular components and supplying metabolic substrates.27 We found that all 7 BCa cell lines showed a certain level of autophagy (Figure 1F). Future studies are needed to elucidate the role of p62-mediated autophagy in BCa.
The impact of NRF2 on cancer is complex. An emerging concept is that NRF2 has its yin and yang side in tumor development.16, 28 On one hand, NRF2 is required for the prevention of chemical and radiation-induced carcinogenesis by quenching ROS and repairing oxidative damage through the expression of its target genes.29-32 On the other hand, recent studies have described that NRF2 activation promotes cancer progression14, 33-35 and metastasis.36 In addition to the genes related to antioxidant response, NRF2 also targets the genes that control proliferation (such as Notch1, Igf1, and Itgb2) and angiogenesis (such as Pdgfc and Vegfc).37 In addition, NRF2 is the key transcription factor that mediates metabolic reprogramming in cancer cells by increasing glucose uptake and directing it to the pentose phosphate pathway.38 Many studies have shown that NRF2 also regulates the tumor microenvironment by reducing the expression of proinflammatory cytokines.16, 39
In summary, we showed that p62 is often overexpressed in human BCa tissue and cell lines. Functionally, p62 promotes BCa cell growth in vitro and in vivo. Mechanically, p62 activates the NRF2-dependent antioxidative response and protects tumor cells from oxidative stress. Our study thus indicated that p62 might serve as a potential therapeutic target in BCa.
DISCLOSURE
The authors declare that they have no conflict of interests.




