RNA-binding protein Musashi2 stabilizing androgen receptor drives prostate cancer progression
Abstract
The androgen receptor (AR) pathway is critical for prostate cancer carcinogenesis and development; however, after 18-24 months of AR blocking therapy, patients invariably progress to castration-resistant prostate cancer (CRPC), which remains an urgent problem to be solved. Therefore, finding key molecules that interact with AR as novel strategies to treat prostate cancer and even CRPC is desperately needed. In the current study, we focused on the regulation of RNA-binding proteins (RBPs) associated with AR and determined that the mRNA and protein levels of AR were highly correlated with Musashi2 (MSI2) levels. MSI2 was upregulated in prostate cancer specimens and significantly correlated with advanced tumor grades. Downregulation of MSI2 in both androgen sensitive and insensitive prostate cancer cells inhibited tumor formation in vivo and decreased cell growth in vitro, which could be reversed by AR overexpression. Mechanistically, MSI2 directly bound to the 3′-untranslated region (UTR) of AR mRNA to increase its stability and, thus, enhanced its transcriptional activity. Our findings illustrate a previously unknown regulatory mechanism in prostate cancer cell proliferation regulated by the MSI2-AR axis and provide novel evidence towards a strategy against prostate cancer.
1 INTRODUCTION
Prostate cancer is one of the most common cancers worldwide and the second leading cause of cancer-related mortality in American males.1 Androgen receptor (AR) plays a key role during prostate carcinogenesis and progression. Once bound and stimulated by androgens, AR is translocated into the nucleus and then activates downstream genes to drive cell growth and proliferation.2 Hence, androgen deprivation therapy (ADT) has become the standard treatment for advanced, relapsed and metastatic prostate cancer and works effectively at first. However, resistance gradually develops, with prostate cancer cells persisting under castration conditions. Almost all patients will eventually progress to the stage referred to as castration-resistant prostate cancer (CRPC), with an average overall survival of 1.5 years.3-5
During the past decade, the mechanism and treatment of CRPC have been a research hotspot. Numerous studies have shown that despite systemic androgen depletion, CRPC continues to be sensitive to AR pathway inhibition, which has highlighted the role of AR in the development of CRPC.5-7 However, resistance against novel ADT, such as abiraterone and enzalutamide therapy, gradually emerges, and this disease remains incurable, with significant morbidity and mortality.8
Posttranscriptional regulation of AR plays an important role in prostate cancer progression. Among posttranscriptional regulators, RNA-binding proteins (RBP) are the master regulators of mRNA processing and translation, regulating RNA splicing, polyadenylation, stability, translation and degradation.9, 10 To date, a variety of RBP have been reported to be involved in the regulation of prostate cancer pathogenesis or progression, which has become a new hotspot for research.11-14 Furthermore, studies have begun to focus on RBPs that participate in AR mRNA stability or splicing. Sam68, which is overexpressed in clinical prostate cancer, controls expression of AR exon 3b to increase endogenous AR-V7 mRNA.15 PSF can induce various AR spliceosome genes and promote production of AR and its variants at the mRNA level in hormone-refractory prostate cancer.16 In addition, heterogeneous nuclear ribonucleoprotein family members, such as HNRNPH1 and HNRNPL, cooperate in the splicing event of AR in CRPC development, contributing to cancer progression.17, 18 However, novel RBP, with the ability to regulate AR mRNA levels implicated in prostate cancer development and even the CRPC stage, remain far from sufficient.
First, we concentrated on RBPs listed in published RBP studies.9, 10 Only 12 RBPs were selected with evident expression differences and then were further analyzed to determine the expression correlation with AR. Second, we discovered a significant correlation between MSI2 and AR expression. Finally, we focused on MSI2, a member of the evolutionarily conserved Musashi RBP family.19 Musashi has two N-terminal RNA recognition motifs (RRM), RRM1 and RRM2, that mediate the binding to motifs located at the 3′-UTR of target mRNA.20 MSI2 has been reported to act as a potent cooperative oncoprotein in hematopoietic malignancies and a variety of solid tumors.21-32
Our study focused on the posttranscriptional regulation of AR by RBPs, and first identified the relationship between MSI2 and AR. Then we demonstrated the high expression of MSI2 in prostate cancer and inhibition of the proliferation of prostate cancer, even CRPC, with MSI2 deletion. Knockdown of MSI2 was accompanied by significantly lower mRNA transcription and protein expression of AR and downstream molecules. Mechanistically, MSI2 binds the 3′-UTR of AR mRNA, and increases AR mRNA stability. Therefore, MSI2 represents an attractive therapeutic target, raising the possibility that MSI2 inhibition could serve as an ADT strategy toward both ASPC and CRPC.
2 MATERIALS AND METHODS
2.1 Cell culture and stable cell line generation
LNCaP, C4-2B, 22RV1, PC3, DU145 and HEK293T cell lines were obtained from ATCC (VA, USA). The normal human prostate epithelial cell line RWPE was stored at the Institute of Urology, Shanghai Jiao Tong University. LNCaP, C4-2B and 22RV1 were used for phenotype and mechanism studies and were authenticated (DOC. S1–S3). Cells were maintained in the appropriate medium (RPMI-1640 for RWPE, LNCaP, C4-2B,22RV1, PC3 and DU145; DMEM for HEK293T) supplemented with 10% FBS, 1% penicillin/streptomycin and were cultured at 37°C with 5% CO2. We generated MSI2 stable knockdown cell lines, using produced lentivirus particles in HEK293T cells which were transfected with a recombinant lentiviral vector containing the shMSI2-1 sequence (5′-GTGGAAGATGTAAAGCAATAT-3′) and shMSI2-2 sequence (5′-CCCAACTTCGTGGCGA CCTAT-3′) or the shCon sequence together with packaging plasmids. In the rescue experiment, siRNA against AR (5′-CUGCUACUCUUCAGCAUUATT-3′) was transfected into the cells using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA).
2.2 Cell proliferation, migration and invasion assay
According to the manufacturer’s instructions for Cell Counting Kit 8 (Dojindo, Kumamoto, Japan), stably transfected cells were processed for cell growth assays (n = 6). For the colony forming experiment, cells were plated in 6-well plates at a density of 2000 cells per well and cultured for 21 days before staining with 0.5% crystal violet (Sangon Biotech, Beijing, China). Clones were calculated by ImageJ software.
The migration and invasion assays were performed by plating 106 cells per well into a 12-well Transwell chamber (Costar) in serum-free medium with or without 1% Matrigel (Corning). Normal growth medium with 10% FBS was placed in the lower chamber. After 24 h, cells in the upper chambers were completely removed with a cotton swab, and the migrated cells attached to the bottom side were fixed in 100% methanol and stained with crystal violet.
2.3 Quantitative real-time PCR
Total RNA was extracted from the indicated cell lines or prostate cancer samples using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. RT-PCR was carried out using PrimeScript RT Master Mix (Takara). Quantitative RT-PCR (qRT-PCR) was performed using SYBR Premix Ex Taq (Takara) in a total volume of 10 μL. The fold change in gene expression was presented using the 2–ΔΔCt method. Experiments were repeated at least twice in triplicate. qPCR primers were as follows: AR, forward 5′-CCTGGCTTCCGCAACTTACAC −3′, reverse 5′-GGACTTGTGCATGCGGTACTCA-3′; MSI2, forward 5′-ATCCCACTACGAAACGCTCC-3′, reverse 5′-GGGGTCAATCGTCTTGGAATC −3′; GAPDH forward 5′-GCACCGTCAAGGCTGAGAAC-3′, reverse 5′-TGGTGAAGACGCCAGTGGA-3′; and M15 forward 5′-TGAAGGCAGA AAAATCCACA-3′, reverse 5′-GTCTGAAGAGGAGTAACTAA-3′. All experiments were performed in triplicate.
2.4 Antibodies, western blotting and coimmunoprecipitation
Whole-cell lysates were prepared with RIPA lysis buffer (Beyotime, Suzhou, China) supplemented with a proteinase inhibitor cocktail (Roche, Basel, Switzerland). A Pierce BCA Protein Assay Kit (Pierce, Rockford, USA) was used to measure protein concentrations. Protein lysates were then separated by SDS-polyacrylamide gel electrophoresis, and target proteins were detected by western blotting analysis with antibodies. The supernatants were immunoprecipitated with 1 µg of the indicated antibodies for 3 h at 4°C, followed by adding 10 µL of protein A/G Plus Agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for a 1 h incubation at 4°C. The immunocomplexes were then washed six times with RIPA buffer containing 5% Tween and examined by western blotting.
The sources of antibodies were as follows: anti–MSI2 (ab76148), anti–AR (ab108341) (Abcam); anti–PSA (2475S) and anti–NKX3.1 (92998S) (Cell Signaling Technology), anti–β-actin (KM9001L) and anti–β-tubulin (KM9003L) (Sungene Biotech).
2.5 Plasmids and dual-luciferase assay
Wild type and mutants (TTAG mutated to AAAA) of 20 potential binding sites in the AR 3′-UTR were cloned into the pmirGLO vector (Promega). FLAG-MSI2 or FLAG-ctrl plasmid and WT or Mut pmirGLO dual-luciferase reporter plasmid were cotransfected into HEK293T cells using Lipofectamine 3000 Reagent. After 48 h, the cells were lysed with 1 × reporter lysis buffer and firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. All experiments were performed in triplicate.
2.6 RNA-immunoprecipitation
C4-2B cells overexpressing FLAG-MSI2 were used for RNA-immunoprecipitation (RIP) with a Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA). Anti–MSI2 was used for protein immunoprecipitation and the RIP experiments were carried out according to the manufacturer’s instructions. Finally, RNA was extracted using the phenol/chloroform method and RNA binding to the MSI2 protein was checked by qRT–PCR. In the site confirmation test, we cotransfected a FLAG-MSI2 plasmid and a Myc-M15 sequence-containing plasmid into HEK293T cells and performed a RIP test to verify the MSI2-M15 binding relationship.
2.7 mRNA decay test and protein stability test
For the mRNA decay test, after transcription inhibition by actinomycin D with gradient time, the AR mRNA abundance in MSI2 stable cell lines was examined by qRT-PCR. For the protein stability test, after stimulation by dihydrotestosterone (DHT) for 24 h, stable cell lines were treated with cycloheximide (CHX) for 0, 1 h, 3 h, 6 h, 12 h or 24 h. Then western blotting was used to show the protein stabilities of AR and its downstream molecules.
2.8 Tissue microarray, immunohistochemistry and immunofluorescence
The prostate cancer tissue microarray contained 61 prostate cancer tissue spots and 39 para cancer spots of 64 patients (OUTDO, Shanghai, China). immunohistochemistry (IHC) and immunofluorescence (IF) were performed as described previously.33 Briefly, IHC was performed on the paraffin embedded tissue microarray using anti–MSI2 (1:200). IHC images were captured by a Leica DM5500 B microscope. Protein expression levels were scored by multiplying the percentage of positive cells and immunostaining intensity. The percentage was scored as follows: nonpositive cells as 0 points, 1%-25% as 1 point, 26%-50% as 2 points, 51%-75% as 3 points and 76%-100% as 4 points. The staining intensity was scored as follows: no positive staining as 0 points, weak staining as 1 point, moderate staining as 2 points and strong staining as 3 points. The final scores were obtained according to above terms: 0-3 was weak expression, 4-8 was moderate expression, and 9-12 was high expression.
Immunofluorescence was performed using anti–human MSI2 (2:100, ab76148, Abcam) and anti–human AR (1:200, NBP2-44789, Novus). IF images of tissues were acquired using a Leica DMi8 fluorescence microscope. Correlation rates were calculated using ImageJ software.
2.9 RNA pulldown
This experiment was previously performed as described34 (AR M15, forward 5′-TCTGTTAAAACTTGTCAGAG-3′, reverse 5′-ATAAATCAAAGCTGAGTCAA-3′; AR M13, forward 5′-CCAAAGCACCAGATCAAATC-3′, reverse 5′-CAAAGAGGCACTAATGCTTG-3′; and FL, forward 5′-CCTGTATTTTCAGGGCATGTCGGAGTGGGACAACATTGCC-3′, reverse 5′-GTACAAGAAAGCTGGGTAAATGCAGATGCAGCATTTATACC-3′). Synthesized RNA were labeled, and the RNA pulldown assay was conducted with the Pierce Magnetic RNA-Protein Pull-Down Kit following the manufacturer’s instructions (Thermo Scientific, Waltham, MA, USA). The result was detected by western blotting.
2.10 Prostate samples
Eight pairs of prostate samples were obtained by radical prostatectomy at Shanghai General Hospital (Shanghai, China) and confirmed by pathology. Approval was obtained from the institutional review board of the Shanghai General Hospital Ethics Committee and written informed consent was obtained from all patients.
2.11 Tumor xenografts
A total of 5 × 106 cells resuspended in 200 μL PBS 1:1 diluted Matrigel were subcutaneously injected in the dorsal flanks of 6-to-8-week-old male nude mice (Animal Center of the Chinese Academy of Sciences, Shanghai, China). Tumor volumes were measured approximately every 3 days and calculated according to the following formula: volume (mm3) = 1/2 × length × width2. Twenty-six days later, the mice were killed and sub-skin grafts were collected for western blotting. The animal experiments were performed in accordance with the US Public Health Service Policy on Humane Care and Use of Laboratory Animals, and were approved by the Scientific Investigation Board of Shanghai General Hospital.
2.12 Statistical analysis
The data are presented as the mean ± SD. Statistical analyses were performed with a two-tailed paired t-test, Fisher’s exact test by using SPSS 17.0 software or GraphPad Prism 6 software. P < 0.05 was considered statistically significant. Pearson’s correlation coefficients were calculated using the ggplot2 package in R version 3.5.0.
3 RESULTS
3.1 MSI2 expression is correlated with androgen receptor expression in prostate cancer
We concentrated on previously reported RBPs9, 10 and analyzed their expression differences between normal prostate tissue and prostate cancer using the published sequence data in the TCGA and GTEx databases. Only 12 RBPs were selected with evident expression differences (Figure S1) and then were further analyzed to find the expression correlation with AR using the ggplot2 package in R version 3.5.0. Interestingly, we found that AR expression was highly correlated with MSI2 expression (r = 0.88, P < 0.05) (Figure S2). Other than AR, we found that MSI2 was highly correlated with AR downstream target genes, such as NKX3.1, KLK3 and TMPRSS2 (Figure S3). Correlations between MSI2 and AR target genes expression in 623 patients with prostate cancer (TCGA database)35 were also investigated (Figure 1A). Pearson’s correlation coefficients were used for the estimation of correlated genes. A total of 755 target genes of the AR transcription factor in low-throughput or high-throughput transcription factor functional studies were from the TRANSFAC Curated Transcription Factor Targets dataset. An IF assay of prostate cancer tissue microarray (Table S1) was performed to further validate the correlation between MSI2 and AR. (r = 0.61, P < 0.05, Figure 1B). Western blotting analysis of eight randomly selected tumor/para-tumor pairs (detailed information is shown in Table S2) and six prostate cell lines (RWPE as the normal human prostate epithelial cell line; LNCaP as the androgen sensitive prostate cell line; and C4-2B, 22RV1, PC3 and DU145 as the androgen-insensitive prostate cancer cell lines) also verified the correlation (r = 0.75, P < 0.05, Figure 1C; r = 0.69, P < 0.05, Figure 1D). These results indicate that MSI2 may be associated with the expression and function of AR in prostate cancer.

3.2 MSI2 expression is upregulated in human prostate cancer and correlated with Gleason scores
Two independent Gene Expression Omnibus (GEO) files (GSE3325 and GSE6919)36, 37 showed obviously higher expression levels of MSI2 in metastatic prostate cancer compared with normal tissue or localized tumor tissue (Figure S4A,B). We searched the data on CRPC in the Oncomine database and then focused on Tomlins Prostate Statistics. Although only two samples were in the hormone-naive group and there was no statistical difference in the data, the result suggested that the median expression level of MSI2 in hormone-refractory patients was higher than that in hormone-naive patients (Figure S4C, P = .83). To evaluate the expression level of MSI2 and its correlation with prostate cancer development, we performed IHC staining using a similar tissue chip as for the IF test. Strikingly, MSI2 was found to be significantly upregulated in tumors compared to para-tumor tissues (P = 0.0001) (Table S3, Figure 2A). Moreover, MSI2 expression was higher in the samples with Gleason score (GS) ≥ 7 compared to those with GS < 7 (P = 0.001) (Figure 2B). Accordingly, western blotting analysis of eight randomly selected para-tumor/tumor pairs and six prostate cell lines confirmed that MSI2 expression was upregulated in prostate cancer samples as well as cell lines (Figure 1C,D).

3.3 MSI2 is required for prostate cancer cell growth in vitro and in vivo
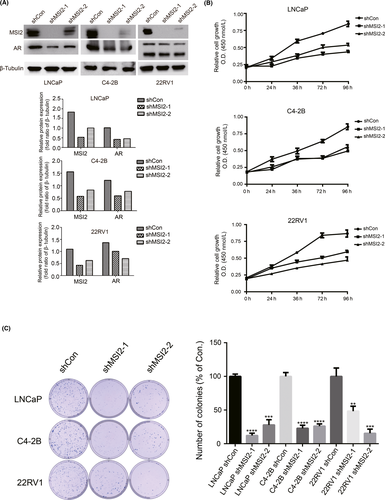
We constructed stable cell lines of LNCaP, C4-2B and 22RV1 cells with stable depletion of MSI2 using lentivirus-mediated shRNA (shMSI2). Western blotting and qRT-PCR results showed significantly lower AR protein expression as well as mRNA transcription in shMSI2 clones (Figures 3A and S5A-C). Compared to that of shRNA-transduced control (shCon) cells, the proliferation rate of shMSI2 clones was significantly reduced, as demonstrated by the CCK-8 assay (Figure 3B). Colony formation assays further showed that MSI2 knockdown reduced the number of colonies formed (Figure 3C).
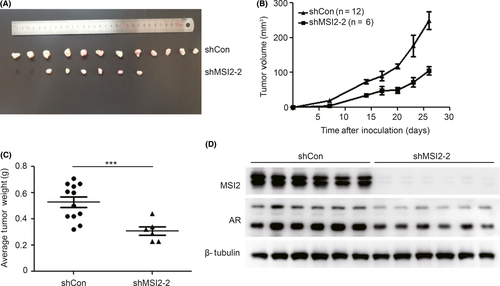
Moreover, to investigate the role of MSI2 in prostate cancer in vivo, we injected stable knockdown 22RV1/shMSI2-2 cells or shCon cells into nude mice to establish tumor xenografts. Twenty nude mice in each group (shCon group and shMSI2-2 group) were injected using stable knockdown 22RV1/shMSI2-2 cells or shCon cells to establish tumor xenografts and were killed on the 26th day. The tumor xenograft incidence rates were different, with 60% (12 in 20) in the shCon group and 30% (6 in 20) in the shMSI2-2 group (Figure 4A), indicating tumorigenesis ability differences between the two stable cell lines. The growth curves suggested that the shMSI2-2 group tumors grew slower than the shCon group (Figure 4B). Similarly, the shMSI2-2 group showed a lower average tumor weight than the shCon group (Figure 4C). MSI2 protein expression in six pairs of xenograft tumors was determined by western blotting (Figure 4D). Accordingly, the significant differences in the incidence rate, growth rate and tumor weight of 22RV1 xenografts suggest that MSI2 is important for prostate cancer xenograft tumor establishment and growth.
To further investigate whether the cell growth regulation of MSI2 was AR-dependent, rescue experiments were performed. These results suggested that overexpression of AR could rescue the proliferation inhibition by MSI2 knockdown (Figure S6). Because AR is a transcriptional factor, we examined MSI2 expression after silencing AR expression in prostate cancer cells but found no change (Figure S7). Furthermore, shMSI2 clones showed inhibited migration and invasion (Figure S8). Collectively, these data suggest that MSI2 plays an important role in regulating AR-dependent growth in prostate cancer cells.
3.4 MSI2 regulates androgen receptor stability and function in prostate cancer cells
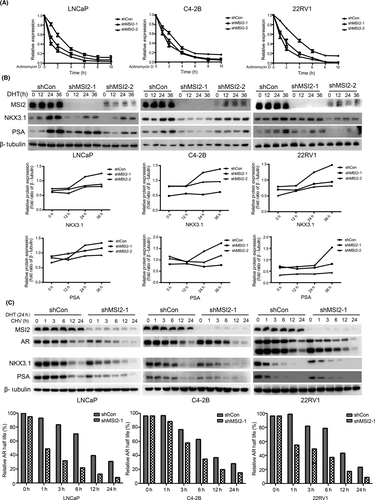
We found inhibited expression of AR in MSI2-knockdown prostate cancer cells, and further studies were carried out to investigate whether MSI2 knockdown could influence AR activity. After transcription inhibition by actinomycin D over a period of time (0, 2, 4, 6, 8 h), the AR mRNA abundance was examined by qRT-PCR. MSI2 knockdown cell lines showed more accelerated mRNA decay than shCon cells (Figure 5A). Upon stimulation by DHT over gradient time (0, 12, 24, 36 h), LNCaP, C4-2B and 22RV1 cells showed similar lower PSA and NKX3.1 protein expression with MSI2 knockdown (Figure 5B). Interestingly, we also found the regulation function of MSI2 in AR protein stability. CHX prevents new protein synthesis. After stimulation with DHT for 24 h, stable cell lines were treated with CHX for 0, 1 h, 3 h, 6 h, 12 h or 24 h, and the CHX time-gradient experiments suggested that AR was rapidly degraded in shMSI2 cells compared with that of shCon cells (Figure 5C). Most proteins are degraded in the following three ways: autophagy, lysosomal-mediated and proteasomal-mediated degradation.38 Contrary to autophagy inhibitors 3-methyladenine (3-MA) and lysosome function inhibitor chloroquine, only the proteasome inhibitor MG132 could rescue the AR protein level in C4-2B shMSI2-1 stable lines (Figure S9A-C). However, the coimmunoprecipitation (co-IP) test did not find the direct binding relationship between AR and MSI2 protein, suggesting that the regulation on AR protein stability was not in a direct contact way (Figure S9D).
3.5 MSI2 regulates androgen receptor stability via direct binding with the androgen receptor 3′UTR in prostate cancer cells
Mechanistically, MSI2 has been reported to bind to recognition motifs located at the 3′-UTR of target mRNA and then upregulate or downregulate relevant protein expression.20 We hypothesized that MSI2 may bind to the 3′-UTR of AR mRNA to mediate AR mRNA stability. We expressed a FLAG-tagged MSI2 protein in LNCaP and C4-2B cells and performed a RIP test to clarify the MSI2-AR relationship. The results showed that AR mRNA was among the MSI2-coprecipitated mRNA molecules (Figure 6A), indicating that MSI2 can directly bind AR mRNA to regulate the AR pathway in prostate cancer cells.
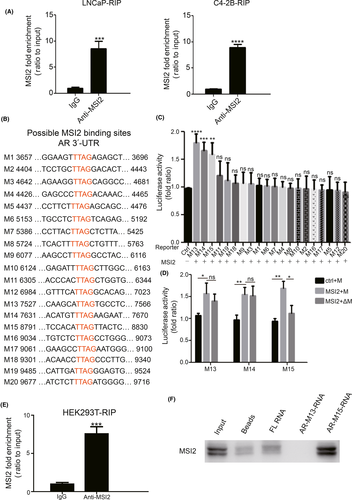
At the molecular level, Musashi proteins were reported to bind r(G/A)U1-3AGU sequences.39 Subsequently, Sujitha’s work identified UUAG as a binding sequence of MSI2.40 We analyzed the AR UTR sequence and found 20 putative binding sites (TTAG; M1-M20) (Figure 6B). Using reporter constructs containing the M1-M20 sites, we performed luciferase assays and the 20 sites were narrowed down to three sites (M13-M15) (Figure 6C). However, when we mutated TTAG to AAAA in these three sites, only the M15 group had statistically significant luciferase activity differences (Figure 6D). Then, we transfected a FLAG-tagged MSI2 plasmid and a Myc-tagged M15 containing sequence plasmid into HEK293T cells and performed a RIP test to verify the MSI2-M15 binding relationship. The results showed that M15 mRNA was coprecipitated with MSI2 (Figure 6E). Finally, we performed an RNA pulldown assay to test the direct association between the M15 site of the AR 3′-UTR and MSI2 (Figure 6F). All these data revealed that MSI2 regulates AR mRNA via direct binding to the M15 site in the 3′-UTR of AR.
4 DISCUSSION
New therapies for prostate cancer are constantly being proposed, such as proapoptotic therapy and immunotherapy, but the effects of these therapies need to be improved. To date, targeting the AR pathway remains the most effective and widely used treatment. Our study first demonstrated the high expression of MSI2 in prostate cancer and its relationship with advanced tumor grade. Then, MSI2 was found to modulate prostate cancer cell growth in vitro and in vivo. Based on the calculated coexpression correlation, we hypothesized that MSI2 might regulate the AR pathway and found that AR overexpression reversed the downregulation of prostate cancer cell proliferation caused by MSI2 knockdown. Mechanistically, the current study determined that MSI2 regulated AR mRNA stability and protein expression, through its direct binding with the 3′UTR of AR mRNA. Collectively, our findings suggest that MSI2 functions as an upregulator of AR both in ASPC and CRPC, and inhibition of MSI2 may contribute to prostate cancer treatment.
Recently, MSI2 has been shown to be a potent cooperative oncogene and found to participate in critical biological processes in human leukemias and some solid tumors. In the current study, we investigated the relationship between MSI2 and prostate cancer development. Bioinformatic analysis, IHC staining and western blotting demonstrated that high expression of MSI2 was frequently detected in prostate cancer tissues and positively correlated with advanced tumor grade. Studies have demonstrated that MSI2 promotes cell growth in breast cancer,41 migration and invasion in bladder cancer42 and drug resistance in pancreatic cancer.43 Similar to the results of previous reports, CCK-8 assays, colony formation assays and xenograft experiments confirmed that MSI2 could enhance prostate cancer cell growth in vitro and in vivo. Transwell analysis also verified the promoting function of MSI2 in prostate cancer migration and invasion. All these results indicate a potential role for MSI2 in a novel, underlying mechanism in the development and progression of prostate cancer.
In 1966, the Nobel Prize was awarded for Huggins’ work showing that prostate cancer was androgen dependent and could be treated by castration.44 The androgen-activated transcription factor AR has been widely investigated. The AR features that contribute to the progression of prostate cancer consist of AR amplification, mutations, variants and alterations of coregulators, all of which lead to AR hyperactivity.5, 45-50 In addition to what we described above, posttranscriptional regulation is also an efficient method for gene regulation. MSI2 is a member of the RBP family, and although MSI2 has been implicated in the development of several solid tumors, the RNA-binding partners of MSI2 have not been completely investigated. For example, overexpressed MSI2 in breast cancer alters the ESR1 by binding specific sites in ESR1 RNA and by increasing ESR1 protein stability.41 Similar to ESR1 in breast cancer, whether the mRNA of AR in prostate cancer could be stabilized by MSI2 was uncertain.
Interestingly, using bioinformatics analysis, western blotting and immunofluorescence, we found that MSI2 was highly correlated with AR and its downstream target genes, indicating that MSI2 may have a functional relationship with AR in prostate cancer. shMSI2 clones showed significantly lower protein expression and mRNA transcription of AR and its downstream molecules. Furthermore, actinomycin D time-course experiments suggested that the mRNA stability of AR was decreased in shMSI2 cells compared with shCon cells.
Mechanistically, Musashi proteins were reported to bind r(G/A)U1-3AGU sequences located at the 3′-UTR of target mRNA then upregulate or downregulate relevant protein expression.20, 39 Sujitha’s work verified UUAG as a binding sequence of MSI2.40 Therefore, we hypothesized that MSI2 might bind the 3′-UTR of AR mRNA; then RIP assays, binding site luciferase assays and RNA pulldown assays were performed to verify this binding site. Overall, MSI2 binds to the M15 site in the 3′-UTR of AR mRNA to modulate AR stability.
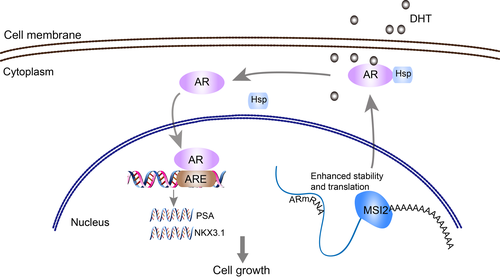
Moreover, the posttranslational regulation of AR to alter AR functional activity, including stability, and cellular localization, also contributes to the progression of prostate cancer.51 To date, few studies have reported the posttranslational regulatory function of RBP, such as the protein-protein interaction between Sam68 and full-length AR or AR-V7.15 We only found two reports indicating a close protein interaction between MSI2 and Numb,52 MDM2 and p5343 in pancreatic cancer. Our CHX time-course experiments showed that AR protein stability was decreased in shMSI2 cells compared with that in shCon cells, indicating a translational regulatory function for AR protein stability of MSI2. The AR protein degradation level is controlled by various pathways, including proteasome, autophagy and lysosome systems.38 While 3-MA and chloroquine could not rescue the AR downregulation caused by MSI2 knockdown, the MG132 inhibitory experiment focused our interest on proteasome-mediated degradation. Then, we supposed that there was a direct binding relationship between AR and MSI2, which was demonstrated not to be the case based on the negative results of co-IP tests. Thus, we regarded MSI2 as an indirect regulator that could modulate AR protein stability through unknown factors. MSI2 upregulates the expression of MDM2, the first E3 ubiquitin ligase identified for the AR, and MSI2 upregulates the PI3K/AKT/mTOR axis, which increases AR degradation in high androgen level environment.43, 53, 54 However, both reported regulatory mechanisms are inconsistent with the upregulated stability in AR protein level caused by MSI2. Our finding, which is encouraging as well as surprising, requires further precise mechanistical studies and will become our next research focus. Overall, our study provides insights into an important role for MSI2 in prostate cancer progression and reveals a novel mechanism by which MSI2 stimulates AR mRNA stability to promote prostate cancer cell proliferation (Figure 7). We found a new treatment target for prostate cancer, and therapy that incorporates its inhibition could contribute significantly to prostate cancer treatment.
ACKNOWLEDGMENTS
We thank Dr Gang Yi and Dr Fuxiang Zhu for technical assistance and helpful discussion. This work was funded by the National Natural Science Foundation of China (No. 81772748 and 81570682) and the Shandong Science and Technology Development Project (2013G0021822).
DISCLOSURE
The authors declare no potential conflicts of interest.




