Serum deprivation response inhibits breast cancer progression by blocking transforming growth factor-β signaling
Funding Information:
National Natural Science Foundation of China (Nos 81372843, 81472472 and 81502518), the National Science and Technology Support Program (No. 2015BAI12B15).
Abstract
Serum deprivation response (SDPR), a key substrate for protein kinase C, play a critical role in inducing membrane curvature and participate in the formation of caveolae. However, the function of SDPR in cancer development and progression is still not clear. Here, we found that SDPR is downregulated in human breast cancer. Overexpression of SDPR suppresses cell proliferation and invasion in MDA-MB-231 cells, while depletion of SDPR promotes cell proliferation and invasion in MCF10A cells. Subsequently, SDPR depletion induces epithelial–mesenchymal transition (EMT)-like phenotype. Finally, knockdown of SDPR activates transforming growth factor-β (TGF-β) signaling by upregulation of TGF-β1 expression. In conclusion, our results showed that SDPR inhibits breast cancer progression by blocking TGF-β signaling.
Serum deprivation response suppresses cell proliferation and invasion in breast cancer cells. SDPR depletion induces epithelial–mesenchymal transition by activation of TGF-β signaling.
Breast cancer is one of the most common malignancies in women worldwide and the second leading cause of cancer-related diseases in women, especially middle and old aged women, the incidence rate of which is still very high.1, 2 Although improvements in early detection and treatment have decreased breast cancer mortality rates in recent years, prevention and therapy of breast cancer remain a major public health concern. Unfortunately, the incomplete understanding of its carcinogenic mechanisms leads to difficulties in selecting targeted treatment and contributes to a low survival rate for patients with breast cancer.
Serum deprivation response factor (SDPR), also known as Cavin-2, is located on chr 2, q32-33, with the function of coding purified phospholipids binding protein of platelets. SDPR has been considered as a key substrate for protein kinase C (PKC) phosphorylation and this interaction determines the compartimentalization of PKC to caveolae.3, 4 SDPR was further confirmed to function in inducing membrane curvature and participate in the caveolae formation.5, 6 Caveolae is calcium channel related to gut electrophysiological pacing function and impacts the cell proliferation and migration.7 Previous studies indicated that SDPR was significantly reduced in tumors of breast, kidney and prostate.8-10 Recently, it has been demonstrated that downregulation of miR-206 induces the deformation of caveolae and suppresses cell proliferation and migration by upregulating SDPR expression.11 These studies suggest that SDPR is a potential tumor suppressor. However, the mechanism of SDPR in breast cancer development and progression is still not clear.
In the present study, we investigated the function of SDPR in breast cancer progression. We found that the downregulation of SDPR expression promotes breast cancer progression by induction of EMT through activation of TGF-β signaling.
Materials and Methods
Reagents and antibodies
Recombinant human TGF-β1 was purchased from R&D System. TGF-β inhibitor SB-431542 was purchased from Selleck (Shanghai, China). The antibodies for SDPR (ab103230) and N-cadherin (ab18203) were purchased from Abcam (Cambridge, MA, USA). The antibodies for pSmad2/3 (9510), Smad2/3 (total; 8685), Cyclin D1 (2926), p21 (2946) and β-catenin (8480) were purchased from Cell Signaling (Beverly, MA, USA). The antibodies for Vimentin (sc-373717), E-cadherin (sc-7870) and β-actin (sc-47778) were purchased from Santa Cruz (Santa Cruz, CA, USA).
Cell culture and tissue specimens
MDA-MB-231, MCF7, T47D, MDA-MB-468, SKBR3 and MCF10A cell lines were obtained from the Type Culture Collection of the Chinese Academy of Sciences. MDA-MB-231 and SKBR3 cell lines were cultured in RPMI-1640 medium (Gibco, Gradn Island, NY, USA) with 10% FBS (Gibco). MFC7, T47D and MDA-MB-468 cell lines were cultured in DMEM medium (Gibco) with 10% FBS. While MCF10A cell lines were supplemented with DMEM (Gibco) with 5% horse serum (Life Technologies, Gradn Island, NY, USA), 10 μg/mL insulin (Sigma, St Louis, MO, USA), 0.5 μg/mL hydrocortisone (Sigma), 20 ng/mL EGF (R&D Systems, Redmond, WA, USA) and 100 ng/mL cholera toxin (Sigma). All the cells were supplemented with 1% penicillin/streptomycin (Gibco), in a 5% CO2 and humidified atmosphere at 37°C.
Cancer specimens were obtained from Tianjin Medical University Cancer Institute and Hospital. Thirty cases of primary breast cancer tissue and the paired adjacent normal breast tissue specimens were included in the present study. After mastectomy surgery, the primary cancer tissues and the adjacent normal tissues were flash-frozen in liquid nitrogen and stored at −80°C. This study was approved by the Institutional Review Board of the Tianjin Medical University Cancer Institute and Hospital and written consent was obtained from all participants.
RNA extraction and reverse transcription quantitative PCR
The total RNA of cultured cells and surgically resected fresh breast tissues were extracted using mirVana PARIS kit (Life Technologies) according to the manufacturer's instructions. Reverse transcription was performed using a First Strand cDNA Synthesis kit (TakaRa, Dalian, China), according to the manufacturer's instructions. The real-time quantitative PCR was performed using GoTaq qPCR Master Mix (Promega, Madison, WI, USA) on a Bio-Rad iQ5 Optical System (Bio-Rad, Berkeley, CA, USA). β-actin was used as an internal control. The primers are listed in the Supplementary material.
siRNA, transfection and generation of stable cell line
siRNA targeting SDPR were bought from Guangzhou RiboBio. The targeting sequences of siRNA are listed in the supplementary material. For transfection, cells were plated at a density of 2 × 105 cells/well in six-well plates. When the cells were 60% confluent, 50 nmol/L siRNA or 4 μg SDPR-HA was transfected into cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer's recommendations. After transfection, the RNA and protein were extracted after 24 and 48 h, respectively. SDPR cDNA was generated from MDA-MB-231 cells, the resultant PCR product of which was connected together with pcDNA3.1 tagged HA (SDPR-HA), and the resulting constructs were confirmed by DNA sequencing. For generation of stable cell lines expressing SDPR-HA, 2 days after transfection, MDA-MB-231 cells were transferred to a 10-cm cell culture dish and selected by medium with 1 mg/mL G418 for approximately 2 weeks. G418 resistant colonies were picked up and identified by western blot.
Cell proliferation assay
For MTT assay, 5 × 103 cells were seeded in 96-well plates per well. Cell viability was examined over the next 5 days. After incubation, the cells were incubated with 20 μL MTT (5 mg/mL in PBS; Sigma) at 37°C for 4 h. Then, the medium was removed and the formazan was dissolved in 150 μL of DMSO (Sigma). The absorbance was measured at 570 nm using a micro-plate auto-reader (Bio-Rad).
For colony formation assay, 24 h after transfection, the cells were seeded into six-well plates at a density of 500 cells/well. After approximately 15 days, the cells grew to visible colonies and were stained with crystal violet. The colonies were counted and compared with control cells.
The EdU assay was detected by EdU labeling/detection kit (Ribobio) according to the manufacturer's protocol. Briefly, after transfection for 48 h, cells were incubated with 25 μM EdU for 12 h. The cells were fixed with 4% formaldehyde for 30 min at room temperature and treated with 0.5% Triton X-100 for 15 min at room temperature for permeabilization. After being washed with PBS, cells were reacted with Apollo reaction cocktail for 30 min before fixation, permeabilization and EdU staining. Subsequently, cell nuclei were stained with Hoechst 33342 at a concentration of 5 μg/mL for 30 min. The cells were then observed under a fluorescence microscope. The percentage of EdU-positive cells was examined by fluorescence microscopy.
Invasion assay
The invasion ability of breast cancer cells in vitro was evaluated by Matrigel coated Transwell inserts (BD Biosciences, San Diego, CA, USA). 1 × 105 cells in 200 μL FBS-free medium were added in the upper chamber of Transwell and 10% FBS-containing medium was added in the lower chamber. After 16 h, the cells were fixed by 4% paraformaldehyde and stained by Giemsa stain (Solarbio, Beijing, China). The cells were then observed under a microscope and the number of migrating cells was counted in five predetermined fields.
Western blot
Cells were lysed in RIPA buffer (containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.25% sodium deoxycholate), 1 mM NaF, 10 mM Na3VO4, 1 mM PMSF and protease inhibitor cocktail (Roche, Indianapolis, IN, USA). After incubation at 4°C for 30 min, the lysate was centrifuged at 15 000 g for 10 min at 4°C. Protein concentration was determined using a BCA assay (Pierce, Rockford, IL, USA). Samples were denatured for 5 min at 95°C and subjected to 10% SDS/PAGE. The separated proteins were transferred to a PVDF membrane (Millipore, Bedford, MA, USA). The membrane was blocked in 5% (w/v) skim milk-TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 8.3) solution, followed by incubation with the primary antibodies diluted in skim milk-TBST solution overnight at 4°C. The membrane was then incubated with the corresponding horseradish peroxidase-conjugated secondary antibody (Cell Signaling) for 1 h at room temperature, and immunoreactive protein bands were visualized by enhanced chemiluminescence reagents (Millipore).
Immunofluorescence assay
About 5 × 104 cells were seeded in a 24-well plate. Cells were washed and fixed in 4% formaldehyde for 15 min at room temperature. The cells were then were permeabilized with 1% Triton X-100 for 15 min at room temperature and blocked with 2% BSA for 30 min at room temperature. Cells were then incubated with primary antibody at 4°C overnight, followed by incubation with FITC-/TRITC-conjugated secondary antibodies for 1 h at room temperature, and then stained with DAPI. Finally, coverslips were observed under a fluorescence microscope.
Flow cytometry analysis
For cell cycle distribution assay, cells were fixed with 70% ethanol at −20°C overnight and washed with PBS. Then, the cells were resuspended in 0.1% Triton-X100/PBS and concomitantly treated with Rnase A and stained with 50 μg/mL propidium iodide (PI) for 15 min. The cell apoptosis assay was assessed by Annexin V Staining Kit (BD Biosciences). Cells were washed twice with PBS and resuspended in the binding buffer, before incubation with Annexin V and PI for 15 min at room temperature. The cell cycle distribution and apoptosis were analyzed using BD FACSCanto II.
TGFβ1 measurement with ELISA and TGFβ signaling assay
Culture supernatants were harvested from treated breast cancer cells, centrifuged to remove cellular debris, and stored at −80°C. TGFβ1 protein levels were measured with a Human TGFβ1 ELISA Kit (Abcam) according to the manufacturer's instructions. Each ELISA was carried out in duplicate for at least three separate experiments. TGFβ signaling activity was determined using Cignal SMAD Reporter Assay Kits (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The SMAD reporter is a mixture of an inducible SMAD-responsive luciferase construct and a constitutively expressing Renilla construct.
Statistical analysis
All the experiments were performed at least twice independently and data presented as mean ± SEM. All statistical analyses were performed using the SPSS 18.0 software system for Windows (SPSS). The statistical significance of difference was calculated using Student's t-test, with significant differences defined as at least a P-value of <0.05.
Results
Serum deprivation response is downregulated in breast cancer
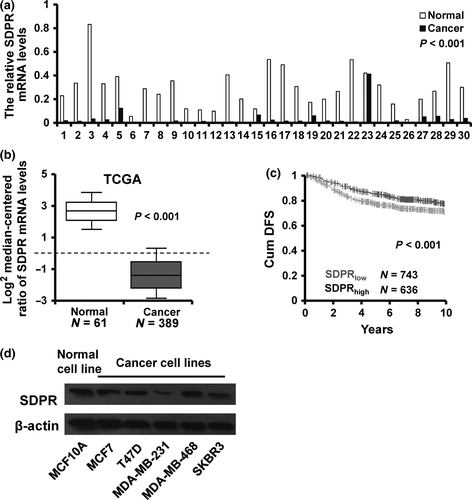
To explore the influence of SDPR on breast cancer progression, we first examined the SDPR mRNA expression in 30 cases of paired normal and breast cancer tissues by reverse transcription quantitative PCR (RT-qPCR). SDPR expression was downregulated in 29/30 collected breast cancer tissues as compared with paired adjacent normal breast tissues (Fig. 1a). To further validate the relationship between SDPR expression and breast cancer progression, we analyzed an SDPR mRNA expression profiling dataset from the Cancer Genome Atlas (TCGA), including 389 cases of breast cancer tissues and 61 cases of normal breast tissues. The validation data confirmed that SDPR mRNA expression was downregulated in breast cancer tissues (Fig. 1b). Moreover, we compared the cumulative disease-free survival (DFS) between patients with high SDPR expression and low SDPR experssion and found that the cumulative DFS with high SDPR expression is higher (red, N = 636) than that of patients with low SDPR expression (gray, N = 743) by using GOBO (Fig. 1c). Next, we assessed the SDPR expression levels in various breast cancer cell lines and normal breast cell line by western blot. The result showed that that the SDPR expression was downregulated in all breast cancer cell lines as compared to MCF10A cells (Fig. 1d). Taken together, these results indicated that SDPR is downregulated in breast cancer.
Serum deprivation response is a tumor suppressor in breast cancer
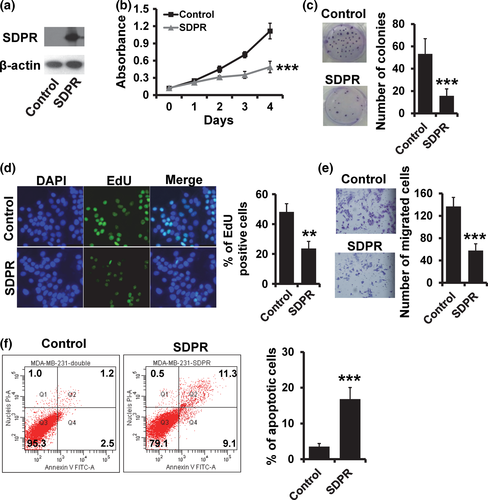
Next, we investigated the influence of SDPR on breast cancer cell proliferation and invasion by generation of stable SDPR-overexpressed MDA-MB-231 cell line (Fig. 2a). Overexpression of SDPR resulted in a lower proliferation rate in MDA-MB-231 cells as assessed by MTT (Fig. 2b), colony formation (Fig. 2c) and EdU (Fig. 2d) assays. Moreover, SDPR overexpression reduced cell invasion in MDA-MB-231 cells (Fig. 2e). We also observed that the apoptotic cells were significantly higher in SDPR-overexpressed MDA-MB-231 cells (Fig. 2f).
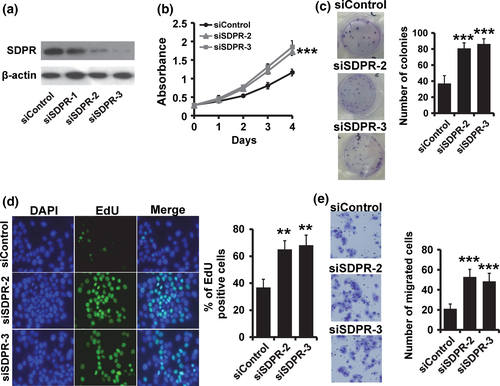
We then examined the effect of SDPR deficiency on the breast cancer cell proliferation and invasion. We used three specific siRNA targeting SDPR, and two of them could efficiently reduce the SDPR expression in MCF10A cells (Fig. 3a). By MTT assay, we found that depletion of SDPR resulted in a substantial increase in the rate of cell proliferation (Fig. 3b). The colony formation assay also showed that depletion of SDPR increased the number of colonies in MCF10A cells (Fig. 3c). Furthermore, the EdU assay confirmed that depletion of SDPR could promote breast cancer cell proliferation (Fig. 3d). The transwell assay showed that depletion of SDPR could promote breast cancer cell invasion (Fig. 3e).
Taken together, these results indicated that depletion of SDPR promotes breast cancer development and progression.
Serum deprivation response suppresses epithelial–mesenchymal transition and blocks cells at G1 phase in breast cancer cells
Epithelial–mesenchymal transition enables epithelial cells to acquire an invasive mesenchymal phenotype and is a critical mechanism for the initial step of metastasis.12 We observed that MDA-MB-231 cells transfected with vector control retained their fibroblast-like morphology, whereas SDPR-overexpressed cells displayed a cobblestone-like morphology (Fig. 4a; upper). Compared with the siControl cells, SDPR depletion transformed MCF10A cells from a typical epithelial morphology into fibroblast-like shape (Fig. 4a; lower). To examine the effect of SDPR expression in breast cancer EMT, we measured the expression of epithelial and mesenchymal markers by RT-qPCR and western blot. SDPR-overexpressed MDA-MB-231 cells exhibited a significant downregulation of vimentin and N-cadherin (CDH2), while the epithelial markers E-cadherin (CDH1) and β-catenin (CTNNB1) were dramatically decreased by RT-qPCR (Fig. 4b) and western blot (Fig. 4d; left). In contrast, the vimentin and CDH2 was upregulated, while the CDH1 and CTNNB1 was downregulated in SDPR-depleted MCF10A cells by RT-qPCR (Fig. 4c) and western blot (Fig. 4d; right). These results showed that SDPR inhibits EMT-like phenotype in breast cancer cells.
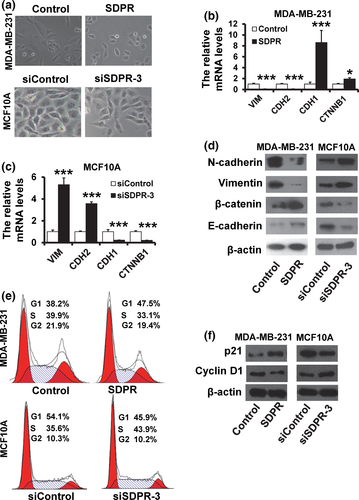
Next, we analyzed the effect of SDPR expression on the cell cycle distribution by flow cytometry analysis. The percentage of cells at G1 phase was increased in SDPR-overexpressed MDA-MB-231 cells (Fig. 4e; upper), whereas the percentage of cells at S phase was increased in SDPR-depleted MCF10A cells (Fig. 4e; lower) compared with those of control cells. Furthermore, SDPR-overexpressed MDA-MB-231 exhibited an upregulation of the cell cycle inhibitor p21 and a downregulation of Cyclin D1. In contrast, p21 was downregulated, while Cyclin D1 was upregulated in SDPR-depleted MCF10A cells (Fig. 4f). These results indicated that SDPR blocks cells at G1 phase in breast cancer cells.
Depletion of Serum deprivation response induces TGF-β signaling
Transforming growth factor-β (TGF-β) is a pleiotropic cytokine which contributes to wound healing, angiogenesis, fibrosis and cancer.12 We observed that the ability of cell proliferation and invasion was reduced after treatment with TGF-β inhibitor SB431542 (Fig. S1a–d). Furthermore, SB431542 could inhibit the EMT-like phenotype in MDA-MB-231 cells (Fig. S1e). To investigate whether TGF-β plays an important role in the function of SDPR, we performed a luciferase assay to determine the effect of SDPR on the TGF-β signaling activity. As shown in Figure 5(a), the luciferase activity was significantly increased in SDPR-depleted MCF10A cells (Fig. 5a). The TGF-β1 expression was also elevated in SDPR-depleted MCF10A cells by ELISA assay (Fig. 5b). The immunofluorescence staining analysis showed that Smad2/3 translocated into the nucleus in SDPR-depleted MCF10A cells more than in the control cells (Fig. 5c). Furthermore, the expression of phosphorylated Smad2/3 were greatly increased in SDPR-depleted cells by western blot (Fig. 5d). The number of proliferating cells was much lower in SDPR-depleted cells after treatment with TGF-β inhibitor SB-431542 (Fig. 5e). Furthermore, the malignant phenotype induced by SDPR depletion was reversed by treatment with TGF-β inhibitor SB-431542 (Fig. 5f–h). We also observed that the percentage of cells at S phase was dramatically decreased in SDPR-depleted MCF10A cells after treatment with SB431542 (Fig. 6i). Furthermore, the expression of p21 was elevated, while the expression of Cyclin D1 was decreased after treatment with SB431542 (Fig. 6j).
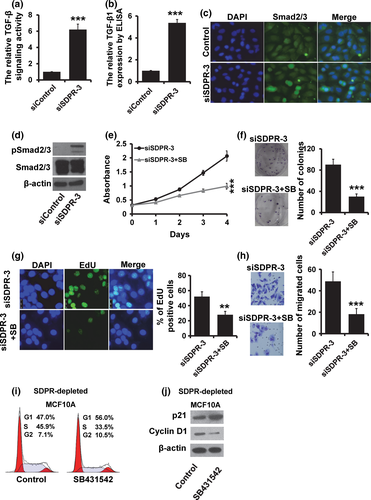
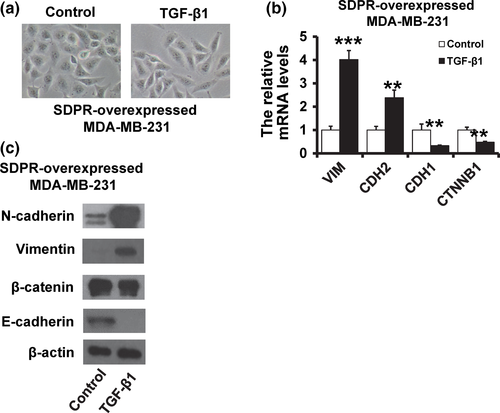
To further confirm that SDPR mediated breast cancer progression through regulating TGF-β signaling, we added TGF-β1 in SDPR-overexpressed MDA-MB-231 cells. We observed that addition of TGF-β1 restored the fibroblast-like morphology (Fig. 6a). Moreover, the expression of epithelial markers was downregulated, while the mesenchymal markers were elevated after treatment with TGF-β1 by RT-qPCR (Fig. 6b) and western blot (Fig. 6c). Together, these results suggested that downregulation of SDPR promotes breast cancer progression through activation of TGF-β signaling.
Discussion
In the present study, we determined the role of SDPR in breast cancer progression. We observed that SDPR expression was downregulated in breast cancer tissues. Overexpression of SDPR supressed breast cancer progression, while depletion of SDPR promoted breast cancer progression. Furthermore, the results showed that SDPR depletion induced EMT and activated TGF-β signaling.
Several reports have suggested that SDPR is most frequently downregulated in cancer tissues, including in breast, kidney and prostate cancer.8-10 Consistent with these studies, our findings demonstrated that SDPR is poorly expressed in breast cancer tissues. Sharan et al. (2015) indicated that SDPR suppresses cell proliferation and migration in 293FT cells.11 Thus, we further assessed whether SDPR suppresses the proliferation and invasion in breast cancer cells. We found that ectopic expression of SDPR could reduce cell proliferation and invasion, and promote cell apoptosis in MDA-MB-231 cells. Moreover, depletion of SDPR enhanced the cell proliferation and invasion in MCF10A cells. Together, these results demonstrated that SDPR suppresses cell proliferation and invasion in breast cancer cells. Based on the gene expression profiling, a molecular classification has been proposed to classify breast cancer into five different subtypes: Luminal A, Luminal B, Basal, HER2+ and normal-like.13 Based on these subtypes, breast cancer patients can be divided into groups with distinct tumor morphologies, prognosis and responses to therapies.14 The role of SDPR in breast cancer differentiation needs to be clarified in future.
Epithelial–mesenchymal transition is a key process underlying tumor cell dissemination from the primary tumor to distant organs. During EMT, epithelial cells lose their cell polarity and molecular expression, enabling cell–cell adhesion, and they gain migratory and invasive properties.15, 16 Lack of SDPR contributes not only to the morphology of the basal plasma membrane, but also to the maintenance of epithelial polarity during early pregnancy.17 In our study, we found that SDPR decreases EMT programs in human breast cancer cells, including upregulation of epithelial markers (E-cadherin and β-catenin) and downregulation of mesenchymal markers (Vimentin and N-cadherin). These findings demonstrated that SDPR inhibits EMT phenotype in breast cancer cells. TGF-β is a multi-functional regulator of cell growth, apoptosis, differentiation and migration.18 Many studies suggest that transforming growth factor-β (TGF-β) can induce EMT.12, 19, 20 TGF-β signaling pathway has been considered as an activator of cancer progression. We observed that depletion of SDPR could elevate the expression of TGF-β1 and activate TGF-β signaling.
In conclusion, we demonstrated that SDPR is downregulated in breast cancer tissues and SDPR supresses breast cancer progression by blocking TGF-β-induced EMT.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos 81372843, 81472472 and 81502518), and the National Science and Technology Support Program (No. 2015BAI12B15).
Disclosure Statement
The authors have no conflict of interest to declare.




