Gene expression profiling of Epstein–Barr virus-positive diffuse large B-cell lymphoma of the elderly reveals alterations of characteristic oncogenetic pathways
Abstract
Epstein–Barr virus (EBV)-positive diffuse large B-cell lymphoma (DLBCL) of the elderly (EBV[+]DLBCL-E) is classified as a subtype of DLBCL. Until now, its molecular pathogenesis has remained unknown. To identify pathways characteristic of EBV(+)DLBCL-E, gene expression profiling of five EBV(+)DLBCL-E and seven EBV-negative DLBCL (EBV[−]DLBCL) cases was undertaken using human oligonucleotide microarray analysis. Gene set enrichment analysis and gene ontology analysis showed that gene sets of the Janus kinase-signal transducer and activator of transcription (JAK-STAT) and nuclear factor kappa B (NF-κB) pathways were enriched in EBV(+)DLBCL-E cases. To confirm the results of the expression profiles, in vitro analysis was performed. Expression profiling analysis showed that high activation of the JAK-STAT and NF-κB pathways was induced by EBV infection into DLBCL cell lines. Activation of the NF-κB pathway was confirmed in EBV-infected cell lines using an electrophoretic mobility shift assay. Western blot analysis revealed an increased protein expression level of phosphorylated signal transducer and activator of transcription 3 (STAT3) in an EBV-infected cell line. Protein expression of phosphorylated STAT3 was frequently observed in lymphoma cells of EBV(+)DLBCL-E clinical samples using immunohistochemistry (EBV[+]DLBCL-E: 80.0% [n = 20/25] versus EBV[−]DLBCL: 38.9% [n = 14/36]; P = 0.001). The results of the present study suggest that activation of the JAK-STAT and NF-κB pathways was characteristic of EBV(+)DLBCL-E, which may reflect the nature of EBV-positive tumor cells. Targeting these pathways as therapies might improve clinical outcomes of EBV(+)DLBCL-E.
Epstein–Barr virus (EBV) is a ubiquitous γ-herpes virus and establishes a lifelong latency in over 90% of the adult population worldwide.1, 2 EBV has a tendency to preferentially infect B lymphocytes.3 Infectious mononucleosis is a disease caused by primary EBV infection in B lymphocytes and occurs mostly in early childhood.4 In vitro analysis shows that EBV transforms human peripheral blood B lymphocytes to generate continuously proliferating lymphoblastoid cell lines. Due to its tendency to infect B lymphocytes, B-cell lymphomas associated with EBV are common and prevalent disorders among malignant lymphomas, which include Burkitt lymphoma, Hodgkin lymphoma, lymphomatoid granulomatosis and lymphoma arising in immunosuppressed patients.5-7
EBV-positive diffuse large B-cell lymphoma (DLBCL) of the elderly (EBV[+]DLBCL-E) is one of the EBV-associated B-cell proliferative disorders. It was first reported in 2003 as a Japanese series involving 22 cases.8 The fourth WHO classification categorizes EBV(+)DLBCL-E as a subtype of DLBCL.9 The definition of EBV(+)DLBCL-E includes the presence of EBV-positive monoclonal B-cell proliferation and usually involves an individual aged over 50 years without any known immunodeficiency or prior lymphoma. An increasing number of reports describe the various clinical features of EBV(+)DLBCL-E.10-17 The reported frequency of EBV(+)DLBCL-E is 5–11% among DLBCL cases.10, 12, 17 The EBV(+)DLBCL-E cases have poor prognosis compared with that of EBV-negative DLBCL (EBV[−]DLBCL) patients.10
Recently, several groups have reported the pathogenesis of EBV(+)DLBCL-E. Dojcinov et al. revealed that EBV(+)DLBCL-E encompasses a wider pathological spectrum than previously recognized.11 The majority of cases of EBV-associated reactive lymphoid hyperplasia comprise a disease with polyclonal B-cell proliferation. In contrast, EBV(+)DLBCL-E cases have exhibited clonal immunoglobulin gene rearrangements in more than half of the cases. Investigators indicated that a reduced number of T-cell repertoires might be the underlying pathogenetic mechanism of EBV(+)DLBCL-E, secondary to immunosenescence.11 Another research group showed that EBV latent membrane protein 1 (LMP1)-positive large-size tumor cells produced CCL17 and CCL22, and the accumulation of CCR4-expressing cells including regulatory T cells was also observed in cases of EBV(+)DLBCL-E.13 However, there are few reports describing gene expression profiling of EBV(+)DLBCL-E compared with EBV(−)DLBCL cases. In the present report, we compared the gene expression profiles of EBV(+)DLBCL-E and EBV(−)DLBCL cases and identified pathways characteristic of EBV(+)DLBCL-E.
Materials and Methods
A detailed description is included in Data S1.
Patient samples and cell lines
We included five EBV(+)DLBCL-E and seven EBV(−)DLBCL cases in the expression profiling analysis (E-MTAB-973). A total of 61 cases (25 cases of EBV[+]DLBCL-E and 36 cases of EBV[−]DLBCL) were included in the immunohistochemical study using the HANS classification18 and additional analysis. Eleven of the 61 cases involved the same patients for which gene expression profiling was conducted. One patient (EBV[+]DLBCL-E_1) who was investigated using microarray analysis did not have available formalin-fixed paraffin-embedded (FFPE) tissue. Patient characteristics are shown in Tables 1 and S1.
| UPN | Sex | Age (years) | Pathological diagnosis | HANS classification | Subtypesa | Extra nodal lesion | Sample analyzed in the present study | Stage | IPI ≧ HI |
|---|---|---|---|---|---|---|---|---|---|
| EBV(−)DLBCL_1 | F | 41 | EBV(−)DLBCL | Non-GCB | ABC | BM, Breast | LN | 4 | Yes |
| EBV(−)DLBCL_2 | F | 48 | EBV(−)DLBCL | GCB | GCB | BM | LN | 4 | No |
| EBV(−)DLBCL_3 | M | 49 | EBV(−)DLBCL | Non-GCB | ABC | Pleural effusion | LN | 4 | Yes |
| EBV(−)DLBCL_4 | M | 52 | EBV(−)DLBCL | Non-GCB | ABC | – | LN | 3 | NA |
| EBV(−)DLBCL_5 | M | 54 | EBV(−)DLBCL | Non-GCB | GCB | BM | LN | 4 | No |
| EBV(−)DLBCL_6 | M | 56 | EBV(−)DLBCL | Non-GCB | ABC | Skin | LN | 4 | Yes |
| EBV(−)DLBCL_7 | M | 67 | EBV(−)DLBCL | GCB | ABC | BM | LN | 4 | Yes |
| EBV(+)DLBCL-E_1 | F | 72 | EBV(+)DLBCL-E | NA | – | BM | LN | 4 | Yes |
| EBV(+)DLBCL-E_2 | M | 62 | EBV(+)DLBCL-E | Non-GCB | – | BM, stomach | LN | 4 | Yes |
| EBV(+)DLBCL-E_3 | M | 60 | EBV(+)DLBCL-E | Non-GCB | – | – | LN | 3 | No |
| EBV(+)DLBCL-E_4 | F | 70 | EBV(+)DLBCL-E | GCB | – | – | LN | 3 | No |
| EBV(+)DLBCL-E_5 | F | 66 | EBV(+)DLBCL-E | Non-GCB | – | BM, liver | BM | 4 | Yes |
- †Subtypes according to gene expression profiling analysis were defined in a previous study.30 −, Negative; +, positive; ABC, activated B-cell; BM, bone marrow; DLBCL, diffuse large B-cell lymphoma; DLBCL-E, diffuse large B-cell lymphoma of the elderly; EBV, Epstein–Barr virus; F, female; GCB, germinal center B-cell; HI, high–intermediate risk; IPI, international prognostic index; LN, lymph nodes; M, male; NA, not available; UPN, unique patient number.
Three germinal center B-cell (GCB)-like DLBCL cell lines (OCI-Ly7, SUDHL6 and SUDHL10), two activated B-cell (ABC)-like DLBCL cell lines (OCI-Ly3 and OCI-Ly10) and a lymphoblastoid cell line (LCL; UH3) were used for the in vitro analysis.
Gene expression profiling and data analysis
Gene expression profiling was performed using the Agilent 44K human oligonucleotide microarray kit (Agilent Technologies, Santa Clara, CA, USA). Data sets using Gene Ontology Analysis (WebGestalt) and Gene Set Enrichment Analysis (GSEA)19 are shown in Table S2.
Data sets of other EBV-related tumors were obtained from the NCBI or ArrayExpress database (GSE17372, GSE13996 and E-MEXP-2996).
In vitro EBV infection
AGS-CR2 cells (a gift from Dr Seiji Maruo, Hokkaido University)20 were infected with enhanced green fluorescent protein (EGFP)-EBV.21 Three EGFP-positive cell lines (SUDHL6, OCI-Ly7 and OCI-Ly3) were successfully obtained.
Western blotting
Phosphorylation of signal transducer and activator of transcription 3 (STAT3) on tyrosine 705 was quantitated with the Stat3 (Tyr705) Antibody Kit (Cell Signaling, Danvers, MA, USA). LMP1 and EBV nuclear antigen 2 (EBNA2) expression of cell lines was confirmed using S12 and PE2 antibodies, respectively.21, 22
Immunohistochemistry
Immunostaining for phosphorylated STAT3 (pSTAT3) (Stat3 [Tyr705] Antibody Kit; Cell Signaling) was performed on FFPE samples according to standard methods. The cut-off value of positive staining was defined as positively stained tumor cells ≥10%, while that of negative staining was <10%.23, 24
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were carried out using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL, USA) according to the manufacturer's protocol, and labeled and unlabeled oligonucleotides specific for nuclear factor kappa B (NF-κB) were synthesized as described previously.25
Results
Genetic characterization of EBV(+)DLBCL-E
After normalization of the raw data, 36 693 probes were used for further analyses. Among these probes, a total of 1432 showed a P > 0.05 with a false discovery rate (FDR) <25%. The number of probes showing high expression in EBV(+)DLBCL-E and EBV(−)DLBCL was 545 (including 474 genes) and 887 (including 767 genes), respectively. First we selected the differentially expressed probes with an average expression level of more than 0.5 log fold-change (268 probes in EBV[+]DLBCL-E and 275 probes in EBV[−]DLBCL), which could separate EBV(+)DLBCL-E cases as a single cluster (Fig. 1).
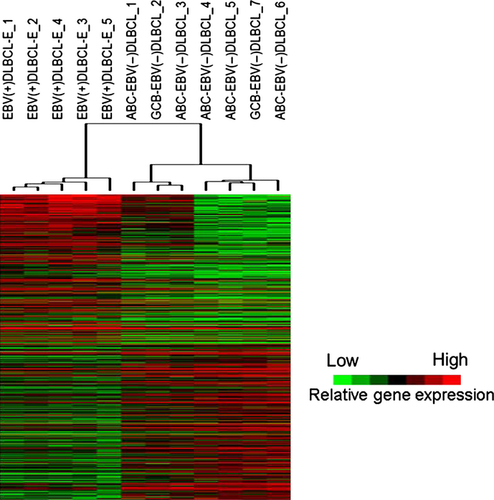
The genes highly expressed in EBV(+)DLBCL-E included immune- and inflammatory-related genes and interleukin-6 (IL6), which were placed in the higher rank. All genes highly expressed in EBV(+)DLBCL-E and EBV(−)DLBCL are shown in Table S3. These results indicate that immune- and inflammatory-related genes are characteristic of EBV(+)DLBCL-E cases.
Pathway analysis revealed Janus kinase-signal transducer and activator of transcription (JAK-STAT) and NF-κB pathway activation in EBV(+)DLBCL-E
For greater characterization, we then performed Gene Ontology Analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the WebGestalt website. The differentially expressed probes with an average expression level of more than 0.5 log fold-change (268 probes in EBV[+]DLBCL-E and 275 probes in EBV[−]DLBCL) were used in both analyses. Additionally, GSEA was carried out using the KEGG pathway-associated gene sets from the Molecular Signatures Database (186 gene sets) (Table S2). These analyses revealed that JAK-STAT, nucleotide-binding oligomerization domain (NOD)-like receptor and Toll-like receptor pathways were characteristic of EBV(+)DLBCL-E. Tables 2, S4 and S5 show the results of the pathway analyses. The protein of STAT3 encoded by this gene is a member of the STAT protein family and the protein of STAT3 is activated through phosphorylation in response to IL6. It is known that STAT3 is activated in B-cell lymphoma. Therefore, we focused on the analysis of STAT3 and performed GSEA analysis using STAT3 and the STAT3 target gene set (Table S2) reported previously23 and found that the gene sets were enriched in EBV(+)DLBCL-E cases (Fig. 2a, Table S6). Constitutive activation of STAT3 was indicated as a characteristic of EBV(+)DLBCL-E.
| Gene ontology category | Genes | P-value |
|---|---|---|
| EBV(+)DLBCL-E | ||
| NOD-like receptor signaling pathway | TNFAIP3, BIRC3, IL1B, IL6, NFKBIA, NOD2 | 6.48E-07 |
| Leishmaniasis | PTGS2, JAK2, IL1A, FOS, IL1B, NFKBIA | 1.21E-06 |
| Osteoclast differentiation | CYLD, IL1A, FOS, IL1B, NFKBIA, SOCS3 | 2.47E-05 |
| Hematopoietic cell lineage | CD55, IL1A, IL1B, IL6, ANPEP | 4.49E-05 |
| JAK-STAT signaling pathway | CCND1, JAK2, IL6, STAT4, IL6ST, SOCS3 | 4.49E-05 |
| Toll-like receptor signaling pathway | LY96, FOS, IL1B, IL6, NFKBIA | 7.47E-05 |
| Pathways in cancer | CCND1, PTGS2, BIRC3, EPAS1, FOS, IL6, NFKBIA | 0.0002 |
| Prion diseases | IL1A, IL1B, IL6 | 0.0003 |
| Apoptosis | BIRC3, IL1A, IL1B, NFKBIA | 0.0003 |
| MAPK signaling pathway | MAP3K5, IL1A, FOS, IL1B, RASGRF2, DUSP1 | 0.0003 |
| EBV(−)DLBCL | ||
| Metabolic pathways | SRM, PIGW, MVK, FASN, CDIPT, POLD1, BCAT2, PDXP, PAFAH1B3, NME7, POLE, GOT2, PYCRL, BDH1, DHFR, SMPD4, DDC, POLD2, EXTL2 | 3.67E-05 |
| Cell cycle | PLK1, ANAPC2, CDC25A, PKMYT1, ESPL1, CDC20, FZR1 | 3.67E-05 |
| Progesterone-mediated oocyte maturation | PLK1, ANAPC2, CDC25A, PKMYT1, AKT1, FZR1 | 4.03E-05 |
| Oocyte meiosis | PLK1, ANAPC2, PKMYT1, SGOL1, ESPL1, CDC20 | 0.0001 |
| Nucleotide excision repair | POLE, GTF2H4, POLD1, POLD2 | 0.0004 |
| Ubiquitin mediated proteolysis | ANAPC2, UBE2S, UBA1, CDC20, FZR1 | 0.0025 |
| Base excision repair | POLE, POLD1, POLD2 | 0.0025 |
| DNA replication | POLE, POLD1, POLD2 | 0.0033 |
| Pyrimidine metabolism | POLE, NME7, POLD1, POLD2 | 0.0054 |
| Arginine and proline metabolism | SRM, GOT2, PYCRL | 0.0084 |
- Epstein-Barr virus (EBV) positive diffuse large B-cell lymphoma (DLBCL) of the elderly [EBV(+)DLBCL-E]; EBV-negative DLBCL [EBV(−)DLBCL]; KEGG, the Kyoto Encyclopedia of Genes and Genomes.
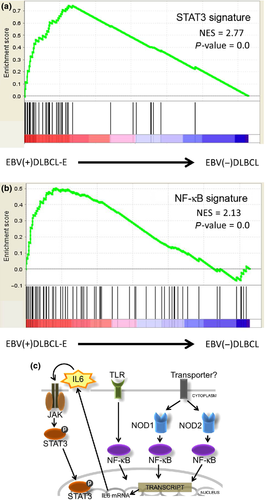
It is known that the JAK-STAT, NOD-like receptor and Toll-like receptor pathways, which were obtained from pathway analysis, are related to the NF-κB pathway26-28 (Fig. 2c). For confirmation, next we performed GSEA using 30 gene sets associated with the NF-κB pathway (Table S2), which were obtained from the GSEA database, and found that eight of the 30 gene sets were enriched (nominal P-val [NOM P-val] < 0.05 and FDR q-val < 0.15) in EBV(+)DLBCL-E compared with EBV(−)DLBCL (Table S6). Furthermore, we evaluated NF-κB pathway activation using a gene set (Table S2) reported by another group29 and found that the gene set was enriched in EBV(+)DLBCL-E cases (Fig. 2b, Table S6). These results suggest that the NF-κB pathway was activated in EBV(+)DLBCL-E cases.
DLBCL is subclassified into two groups according to microarray analysis: GCB-like DLBCL and ABC-like DLBCL. In comparison with GCB-like DLBCL, ABC-like DLBCL has been reported as showing increased activation of the NF-κB pathway.29 We selected ABC-like EBV(−)DLBCL patients among the seven EBV(−)DLBCL cases whose diagnosis regarding GCB-like DLBCL and ABC-like DLBCL was made using expression profiles in our previous study.30 Five of the seven cases were classified as ABC-like DLBCL. We compared five cases of EBV(+)DLBCL-E with five cases of ABC-like EBV(−)DLBCL using the STAT3 and NF-κB gene sets described above and found that both gene sets were enriched in EBV(+)DLBCL-E cases (Table S6). These results indicate that enrichment of STAT3 and NF-κB genes was significant in cases of EBV(+)DLBCL-E.
JAK-STAT and NF-κB pathway activation in EBV-infected B-cell lines
The results of the expression profile suggest that STAT3 and NF-κB activation were characteristics of EBV(+)DLBCL-E. However, it is not clear whether the activation reflected the nature of the tumor cells or the non-tumor cell of the samples, and if STAT3 and NF-κB activation was associated with EBV infection into tumor cells. To investigate the association between EBV-infected B-cell lymphoma cells and molecular pathways, we then conducted in vitro functional analysis. A recombinant EBV harboring EGFP and neomycin resistance genes20 was used to infect five DLBCL cell lines, of which two EBV-infected GCB-like DLBCL cell lines (SUDHL6 and OCI-Ly7) and one EBV-infected ABC-like DLBCL cell line (OCI-Ly3) were successfully established showing ≥90% EGFP positivity (Fig. 3a). The protein expression of LMP1 in SUDHL6 and OCI-Ly3 was confirmed; however, the expression level in OCI-Ly7 was quite low (Figs 3b, S1). SUDHL6 and OCI-Ly7 cell lines were successfully used in studying the protein expression of EBNA2. Expression of EBNA2 was not detected in both cell lines (Fig. S2). Gene expression profiling of EBV-infected cell lines was compared with cell lines before EBV infection using STAT3- and NF-κB-related gene sets (Table S2). Figure 3(c,d) shows that the gene sets of STAT3 and NF-κB were enriched in EBV-infected B-cell lines. To confirm the protein expression level, we performed western blot analysis using the cell lines and found elevation of STAT3 and enhanced pSTAT3 expression after EBV infection into SUDHL6, whereas the expression was not observed in OCI-Ly7 (Fig. 3e). The expression level of pSTAT3 of EBV-infected OCI-Ly3 was the same as that of EBV-naïve OCI-Ly3 (Fig. S1).
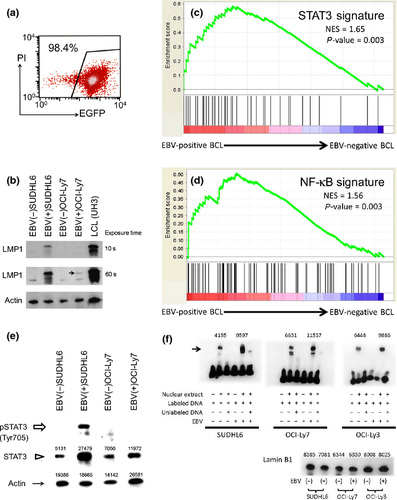
An electrophoretic mobility shift assay was performed to confirm activation of NF-κB. SUDHL-6, OCI-Ly7 and OCI-Ly3 cell lines were infected with EBV as described above. Figure 3(f) shows the results of the electrophoretic mobility shift assay. Each EBV-infected cell line shows increased NF-κB activity compared with the cell line without EBV infection. These results suggest that NF-κB pathway activation was induced by EBV infection into B-cell lymphoma cell lines.
JAK-STAT pathway and NF-κB activation in EBV-related tumors
To investigate the possible effect of the NF-κB pathway and STAT3 activation in other EBV-associated lymphomas, we performed expression analysis using the datasets of several tumors obtained from the database (EBV-positive [n = 9] and EBV-negative [n = 4] human immunodeficiency virus [HIV] DLBCL, GSE17372; EBV-positive [n = 18] and EBV-negative [n = 34] Hodgkin lymphoma, GSE13996; and EBV-positive natural killer [NK] cell lines [n = 7] and normal NK cells [n = 3], E-MEXP-2996). We carried out GSEA using the same STAT3 and NF-κB gene sets (Table S2) and found that NF-κB activation was also observed in other EBV-associated lymphomas (NOM P-val < 0.05; Table S7). Enrichment of STAT3 and the STAT3 target gene set23 was prominent in EBV-positive HIV DLBCL and Hodgkin lymphoma (NOM P-val < 0.05; Table S7). However, the EBV-positive NK cell line did not show high STAT3 activation compared with the normal NK cells (Table S7). These results suggest that STAT3 activation and NF-κB pathway activation are characteristics of EBV-positive B-cell lymphoma.
Immunohistochemical analysis using pSTAT3
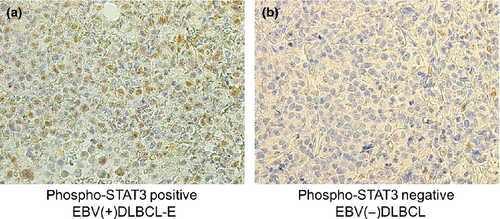
A total of 61 cases (36 cases of EBV[−]DLBCL and 25 cases of EBV[+]DLBCL-E) were analyzed immunohistochemically for pSTAT3 (Table S1). Among the 36 cases of EBV(−)DLBCL, 18 were GCB-like EBV(−)DLBCL and 18 were ABC-like (non-GCB) EBV(−)DLBCL, while the 25 cases of EBV(+)DLBCL-E comprised 22 cases classified as ABC-like (non-GCB) EBV(+)DLBCL-E and three cases classified as GCB-like EBV(+)DLBCL-E using the HANS classification. A total of 20 (80.0%, n = 20/25) EBV(+)DLBCL-E cases showed positivity for pSTAT3 (Fig. 4a). In contrast, positivity was confirmed in 14 cases of EBV(−)DLBCL (38.9%, n = 14/36) (Fig. 4b). The pSTAT3-positive cases of GCB-like and ABC-like (non-GCB) EBV(−)DLBCL included six (33.3%, n = 6/18) and eight (44.4%, n = 8/18) patients, respectively. The high frequency of protein expression of pSTAT3 was confirmed in EBV(+)DLBCL-E cases in comparison with EBV(−)DLBCL (P = 0.001, Student's t-test), which supported the results of the gene expression analysis. Since it is known that ABC-like (non-GCB) EBV(−)DLBCL patients exhibit activation of STAT3,23, 28, 31 we limited analyses to comparison with groups of EBV(−)DLBCL showing the ABC-like (non-GCB) phenotypes. Activation of STAT3 was more prominent in EBV(+)DLBCL-E cases compared with those involving ABC-like (non-GCB) EBV(−)DLBCL patients (P = 0.016, Student's t-test). These results indicate that STAT3 activation is a prominent characteristic of the tumor cells of EBV(+)DLBCL-E.
Discussion
The present study demonstrated that the prominent genes characteristic of EBV(+)DLBCL-E included immune- and inflammatory-related genes. The morphological feature of EBV(+)DLBCL-E exhibited a variable component of reactive elements such as small lymphocytes, plasma cells, histiocytes and epithelioid cells.8, 10, 12 The results of the expression analysis in the present study might reflect the pathological character of EBV(+)DLBCL-E.
There are some reports describing gene expression profiles of EBV-associated B-cell disorders.32, 33 Tousseyn et al. compared EBV-positive post-transplant lymphoproliferative disorder (PTLD) with EBV-negative PTLD using expression profiles and found that immune and inflammatory cytokine-related genes were upregulated in EBV-positive PTLD.32 In EBV-positive PTLD patients, viral-associated genes such as IRF7 (a known regulator of EBV LMP1 expression), EBI1 (EBV-induced gene 1) and interferon-induced genes were overexpressed and the PTLD cells may have originated from memory and activated B-cells.33, 34 EBV/human herpesvirus 8-related HIV lymphoma patients exhibit a high level of serum IL6 and IL10 and disruption of the cytokine network could be a characteristic of these patients.9 Because similar inflammation and cytokine disorders were also observed in EBV(+)DLBCL-E, these imbalances might be common molecular pathogenic features of EBV-associated disorders.
A comparison between EBV(+)DLBCL-E and EBV(−)DLBCL indicated that upregulation of the genes associated with the JAK-STAT and NF-κB pathways was characteristic of EBV(+)DLBCL-E. Activation of the JAK-STAT and NF-κB pathways has been observed in both malignant tumor cells and inflammatory cells infiltrating tumor tissues.35-37 We performed in vitro analyses and immunohistochemistry to determine the components that contribute to activation of the JAK-STAT and NF-κB pathways in EBV(+)DLBCL-E. The JAK-STAT and NF-κB pathway-related genes were upregulated following EBV infection in DLBCL cell lines. Immunohistochemical analysis of EBV(+)DLBCL-E cases revealed that most of the cells positive for pSTAT3 were tumor cells. These results suggest that the EBV-infected tumor cells and not the infiltrating inflammatory cells contribute to activation of the JAK-STAT and NF-κB pathways in EBV(+)DLBCL-E.
It has been reported that ABC-like DLBCL exhibit activation of the JAK-STAT and NF-κB pathways compared with GCB-like DLBCL.23, 28, 31 Therefore, in the present study, we limit analysis to cases involving ABC-like EBV(−)DLBCL patients and EBV(+)DLBCL-E. The results show that the genes associated with activation of the JAK-STAT and NF-κB pathways were more prominent in patients with EBV(+)DLBCL-E compared with ABC-like EBV(−)DLBCL. The result indicates that activation of the JAK-STAT and NF-κB pathways is thought to be characteristic of EBV(+)DLBCL-E. Montes-Moreno et al. reported the pathological findings of 47 cases of EBV(+)DLBCL-E and showed that EBV(+)DLBCL-E is an aggressive B-cell lymphoma with a post-germinal center B-cell phenotype characterized by NF-κB activation.38 Although the present study included a relatively small number of patients, their pathological study supports the validity of our analysis.
Activation of the JAK-STAT and NF-kB pathways is highly regulated in normal cells. In contrast, the LCL comprising transformed B cells is characterized by dysregulation of these pathways.3, 35, 39, 40 LMP1-triggered signaling to JAK-STAT and NF-κB is thought to be initiated at the C-terminal cytoplasmic domain of LMP1, which has a cell-signaling domain in the C-terminal activating region and binds to tumor necrosis factor (TNF) receptor-associated factor, TNF receptor type 1-associated death domain protein and other molecules.3, 35, 39, 40 In cases with EBV-related lymphomas, a 30-bp deleting mutation located in the C-terminal domain of LMP1 has been reported to prolong the half-life of LMP-1, resulting in a higher level of LMP1 expression in cells.41 This mutation could be associated with activation of the NF-κB pathway since LMP1 engages the intracellular CD40 signaling machinery in Hodgkin lymphoma to induce strong constitutive activation of NF-κB,3, 35 and chronic or aberrant STAT activation would be both a necessary and predisposing event for EBV-driven tumorigenesis in immunocompetent individuals.41, 42 In the present study, we used an in vitro model of EBV-infected B-cell lymphoma and found high activation of the pathways was induced in some cell lines. Oncogenic alteration in addition to EBV infection might synergistically contribute to increase the activation of STAT3 and NF-κB in patients with EBV(+)DLBCL-E. Comprehensive genetic studies including LMP-1 and a larger sample series would clarify the pathogenesis of EBV(+)DLBCL-E in the future.
The PI3K/Akt signaling pathway is also known to be activated by LMP1.43 In this analysis, gene expression profiling revealed that the PI3K/Akt pathway was activated in EBV-infected B-cell lines compared with EBV-negative B-cell lines. However, the PI3K/Akt pathway was not activated in clinical samples of EBV(+)DLBCL-E compared with EBV(−)DLBCL (data not shown). Recently, several reports have shown the importance of the tumor microenvironment and its effects on tumor characteristics.44, 45 Although the responses of tumor cells to EBV infection could play an important role in the pathogenesis of EBV(+)DLBCL-E, activation of the signaling pathway might be altered by an interaction between tumor cells and the tumor microenvironment.
In conclusion, the present study identified characteristic oncogenetic pathways when comparing gene expression in EBV(+)DLBCL-E and EBV(−)DLBCL. Activation of JAK-STAT and NF-κB pathways was prominent in EBV(+)DLBCL-E, which may reflect the nature of EBV-positive tumor cells. The therapies targeting these pathways could improve clinical outcomes of some parts of EBV-related B-cell lymphoma. Further studies are needed to investigate the molecular and biological characteristics of EBV(+)DLBCL-E.
Acknowledgements
This work was supported in part by Grants-in-Aid from the Ministry of Health, Labor and Welfare, the Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science (JSPS KAKENHI Grant Number 20591142) and a Grant-in-Aid for Aichi Cancer Center Project Research.
The authors thank Drs Masao Nakagawa, Akira Umino, Keiichiro Honma, Yoshitoyo Kagami, Kiyoko Yamamoto, Tomoko Miyata, Kotaro Arita, Noriaki Yoshida, Naoko Asano, Fang Liu and Eisaku Kondo for their helpful and inspiring discussions, as well as Kyoko Hirano, Yumiko Kasugai, Noriko Saito, Yuko Mizuno and Hiroshi Yamada for their excellent laboratory work.
Disclosure Statement
The authors have no conflict of interest.




