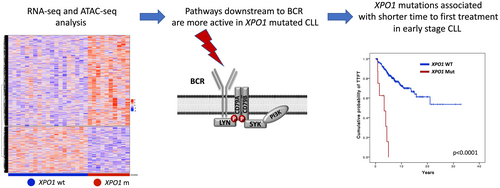
XPO1 mutations identify early-stage CLL characterized by shorter time to first treatment and enhanced BCR signalling
Davide Rossi and Gianluca Gaidano equally contributed to this work.
[Corrections made on 4 October 2023, after first online publication: The 2nd author’s name was corrected and ORCID was added to Gianluca Gaidano in this version.]
Summary
Here we evaluated the epigenomic and transcriptomic profile of XPO1 mutant chronic lymphocytic leukaemia (CLL) and their clinical phenotype. By ATAC-seq, chromatin regions that were more accessible in XPO1 mutated CLL were enriched of binding sites for transcription factors regulated by pathways emanating from the B-cell receptor (BCR), including NF-κB signalling, p38-JNK and RAS-RAF-MEK-ERK. XPO1 mutant CLL, consistent with the chromatin accessibility changes, were enriched with transcriptomic features associated with BCR and cytokine signalling. By combining epigenomic and transcriptomic data, MIR155HG, the host gene of miR-155, and MYB, the transcription factor that positively regulates MIR155HG, were upregulated by RNA-seq and their promoters were more accessible by ATAC-seq. To evaluate the clinical impact of XPO1 mutations, we investigated a total of 957 early-stage CLL subdivided into 3 independent cohorts (N = 276, N = 286 and N = 395). Next-generation sequencing analysis identified XPO1 mutations as a novel predictor of shorter time to first treatment (TTFT) in all cohorts. Notably, XPO1 mutations maintained their prognostic value independent of the immunoglobulin heavy chain variable status and early-stage prognostic models. These data suggest that XPO1 mutations, conceivably through increased miR-155 levels, may enhance BCR signalling leading to higher proliferation and shorter TTFT in early-stage CLL.
Graphical Abstract
INTRODUCTION
Genomic analysis of chronic lymphocytic leukaemia (CLL) has clarified the disease molecular landscape and has identified a panel of driver genes that includes XPO1.1-3 XPO1 codes for exportin-1, which is one of the most important human shuttle proteins regulating the traffic of macromolecules between nucleus and cytoplasm, and is essential for cellular homeostasis.4 XPO1 binds to a diverse array of proteins through their canonical leucine-rich nuclear export signal (NES) domain that serves as a consensus sequence for nuclear export.4 XPO1, in addition to CLL, is mutated in several other haematological and solid tumours.4 Most XPO1 mutations detected in human cancer consist of the E571K substitution that confers a gain-of-function. In cells harbouring XPO1E571K mutations, a negatively charged glutamic acid at position E571 is substituted with a positively charged lysine. This change enhances interaction of XPO1E571K with proteins bearing negatively charged NES sequences thus translating into an augmented export of negatively charged cargo proteins.5
The B-cell receptor (BCR) signalling is essential for CLL pathogenesis and proliferation.6, 7 Compared to normal B cells, the BCR of many CLL cells is characterized by an intrinsically higher reactivity to antigens.6, 7 In cases of unmutated immunoglobulin heavy chain variable (IGHV) rearrangements and in the presence of specific stereotyped BCR (e.g. subset #2), BCR activation is also enhanced in an antigen-independent manner driven by homotypic BCR to BCR interactions.8, 9 Few data are reported about the interplay between specific gene mutations and BCR activity.
Most of newly diagnosed CLL patients with CLL do not require therapy initially, as they present a lymphocytosis but do not have any additional symptoms or complete blood count abnormalities, and do not have enlarged lymph nodes or splenomegaly.10 These asymptomatic patients, namely Binet A and Rai 0 CLL, are managed with a watch-and-wait strategy since early intervention with chemo-immunotherapy and/or pathway inhibitors did not show a survival benefit.10-14 Since most patients do not require therapy at the time of the diagnosis, CLL provides an informative model to evaluate the disease's intrinsic mechanisms of clonal expansion without the external stimuli imposed by therapy.15-17 Two different prognostic models in early-stage CLL patients identified simple biological and clinical variables that clearly segregate patients who require treatment soon after the diagnosis and patients with a high probability to remain asymptomatic for decades.18, 19 Since CLL is characterized by a high grade of molecular heterogeneity, the analysis of gene mutations may further improve the stratification of time to first treatment (TTFT) in asymptomatic CLL patients.20
The aims of the present study were (i) to evaluate the prevalence of XPO1 mutations in different phases of the CLL clinical course; (ii) to characterize the epigenomic and transcriptomic profile of XPO1 mutant versus XPO1 wild-type CLL and (iii) to evaluate whether XPO1 mutations may predispose to disease progression and early treatment requirement.
MATERIALS AND METHODS
Patients and XPO1 analysis
To evaluate the prevalence of XPO1 mutations in different phases of the disease, we screened for XPO1 mutations 95 monoclonal B-cell lymphocytosis (MBL), 957 early stage CLL (Rai 0/I or Binet A), 81 CLL relapsed after chemoimmunotherapy or pathway inhibitors, and 55 CLL transformed into an aggressive lymphoma (Richter syndrome). The clinical correlation between XPO1 mutations and TTFT was analysed in 276 consecutive newly diagnosed Rai 0 and I CLL patients referring to our institution (1st cohort). The 2nd cohort was represented by a multicentre cohort of 286 Binet A CLL and the 3rd cohort by a multicentre cohort of 395 Rai 0 CLL previously described elsewhere.19 To evaluate the potential clonal evolution of XPO1 mutations, we re-analysed patients followed at our institution at the time of treatment requirement (N = 61) and at the time of Richter transformation (N = 29). We also analysed the XPO1 gene in 33 CLL cases during ibrutinib therapy at homogeneous time points every 24 weeks for at least 96 weeks. The study was approved by the local ethical committee (study number CE 120/19).
The XPO1 gene (exons 15 and 16) was analysed by next-generation sequencing using a targeted resequencing gene panel including the coding exons plus splice sites of 10 CLL driver genes (size of the target region: 26680 bp) in the 1st and the 3rd cohorts (Table S1). In the 2nd cohort, the XPO1 gene was analysed by Sanger sequencing (Supplementary Appendix and Table S2). Multiplexed libraries (n = 10 per run) were sequenced on Illumina MiSeq platform. The variant allele frequency (VAF) threshold was 5%. A robust and previously validated bioinformatics pipeline was used for variant calling.21-23 More precisely, the variants called by VarScan 2 were annotated by using the SeattleSeq Annotation 138 tool (http://snp.gs.washington.edu/SeattleSeqAnnotation138) with the default setting. Variants annotated as SNPs according to dbSNP 138 (with the exception of TP53 variants that were manually curated and scored as SNPs according to the International Agency for Research on Cancer TP53 database; http://p53.iarc.fr), intronic variants mapping >2 bp before the start or after the end of coding exons, and synonymous variants were then filtered out. Among the remaining variants, only protein truncating variants (i.e. indels, stop codons and splice site mutations), as well as missense variants not included in the dbSNP 138 and annotated as somatic in the COSMIC v85 database (https://cancer.sanger.ac.uk/cosmic), were retained.
RNA-seq and ATAC-seq on primary CLL cells
Peripheral blood mononuclear cells from eight patients with XPO1 mutations were sorted to purify the CLL CD19+/CD5+ tumoral cells and were subjected to RNAseq and ATACseq. Fifteen XPO1 wild-type cases, matched for IGHV status, TP53 status and FISH karyotype, were analysed for comparative purposes (Table S3). Further information is provided in the Supplementary Appendix.
Assessment of miR-155-5p expression
Cellular RNA was isolated by Trizol (Invitrogen) from 8 XPO1 mutated CLL and from 8 XPO1 wild type CLL matched for IGHV status, TP53 status and FISH karyotype. The cDNA templates were prepared using the TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems). RT-qPCR reactions were performed using the TaqMan Fast Advanced Master Mix (Applied Biosystems) with the miR-155-5p TaqMan Advanced MicroRNA Assay. The miR-186-5p was used as housekeeping gene. Experiments were carried out in duplicate. Quantitative miR expression data were acquired using the StepOnePlus Real-Time PCR system. The relative miR-155-5p expression levels were determined using the comparative Ct (2−ΔΔCt) method and the relative expression between the two groups was compared by the Wilcoxon rank sum exact test.
Statistical methods
The study endpoint was TTFT, defined as the time between presentation and start of first treatment of CLL because of progression to symptomatic disease according to the National Cancer Institute-Working Group/International Workshop on Chronic Lymphocytic Leukemia guidelines (patients without a documented event were censored at the date of last observation or death).10, 24 Survival analysis was performed by the Kaplan–Meier method and compared between strata using the Log-rank test. The adjusted effects of XPO1 mutations and the IPS-E18 and Rai 0 prognostic model19 variables on TTFT were estimated by Cox regression. The analysis was performed with the Statistical Package for the Social Sciences software v.24.0 (Chicago, IL).
RESULTS
Frequency of XPO1 mutations in different phases of CLL
The prevalence of XPO1 mutations was assessed across different clinical phases of CLL, including MBL (n = 95), early-stage CLL (n = 957), CLL relapsed after chemoimmunotherapy or pathway inhibitors (n = 81), and CLL transformed into an aggressive lymphoma (Richter syndrome) (n = 55). XPO1 mutations occurred in 3.1% (n = 3/95) of MBL, 2.9% (n = 28/957) of early-stage CLL, 4.9% (n = 4/81) of CLL relapsed after chemoimmunotherapy or pathway inhibitors and 12.7% (7/55) of CLL transformed into diffuse large B-cell lymphoma (Richter syndrome) (Figure S1A). The percentage of XPO1 mutated patients was significantly higher in Richter syndrome when compared to MBL (12.7% vs. 3.1%, p = 0.03) and early-stage CLL patients (12.7% vs. 2.9%, p value = 0.002), respectively (Figure S1A).
Among Rai 0/I CLL provided with complete clinical and molecular data (n = 276), after adjusting for multiple comparisons, XPO1 mutations significantly correlated with an unmutated IGHV genes status and with a lymphocyte doubling time (LDT) < 6 months (p = 0.02; Figure S2).
Mutations of XPO1 clustered in two hotspots. The first hotspot mapped at p.E571 accounted for 92.8% (39/42) of all mutations and affected a phylogenetically conserved position (Figure S1B). The glutamic acid in position 571 was commonly mutated into lysine or less frequently into glycine, alanine, or valine (Figure S1B). Mutations were selected to change the negatively charged glutamic acid of position 571 into a positively charged amino acid. The second hotspot mapped at p.D624 accounted for 7.1% (3/42) of mutations and appeared to be selectively restricted to CLL, while variants affecting the p.E571 position have been described across different cancer types.5
To evaluate the potential clonal expansion of XPO1 mutations during the CLL clinical course, we screened sequential samples of (i) CLL at the time of treatment requirement (N = 61), (ii) CLL on ibrutinib treatment (N = 33), and (iii) CLL transformed into Richter syndrome (N = 29). The VAF of XPO1 mutations remained stable over time (19.0% at the time of diagnosis vs. 27.0% at the time of treatment requirement, p = 0.206) and no patient acquired XPO1 mutations during the watch & wait period. Similarly, ibrutinib treatment did not associate with the emergence of XPO1 mutations, as documented in 33 CLL cases followed at homogeneous time points every 24 weeks for at least 96 weeks.
Concerning XPO1 mutation acquisition in Richer syndrome, 29 patients were analysed both in the CLL phase and in the lymph node biopsy affected by Richter syndrome. XPO1 mutations were identified in 4 out of 29 cases of clonally related Richter syndrome. Three out of 4 cases displayed a mutation of XPO1 in the Richter lymph node biopsy but not in the matched CLL phase, suggesting that XPO1 mutations had been conceivably acquired at the time of Richter transformation.
XPO1 mutations impact on chromatin accessibility of CLL cells
To gain insights into the proliferative course of XPO1-mutated CLL, we searched for cellular programs associated with XPO1 mutations. CLL cells were purified through flow-sorting from 8 patients harbouring XPO1 mutations (VAF from 25% to 50%) and from 15 patients lacking XPO1 mutations used as controls. XPO1 mutated and XPO1 wild-type CLL samples were matched for the main biological CLL characteristics (IGHV, TP53 status and FISH karyotype) to mitigate biases related to the disease genetic background. Samples were profiled for both chromatin accessibility and transcriptome.
By principal component analysis (PCA), XPO1 mutated CLL clearly separated and showed a distinct chromatin profile compared to wild-type cases (p = 0.00356) (Figure 1A). Overall, XPO1 mutated CLL had a chromatin landscape more accessible than that of XPO1 wild type CLL, with 6391 enhancers/promoters being more accessible and only 3283 enhancers/promoters being less accessible in XPO1 mutated compared to XPO1 wild type (FDR < 0.1).
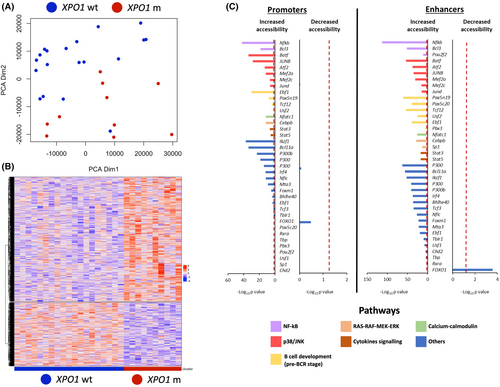
Differentially accessible chromatin regions were decorated with the CLL-specific map of enhancers and active promoters, and with the map of 96 transcription factor binding sites of B cells25 (Figure 1B). Chromatin regions that were more accessible in XPO1 mutated CLL were enriched in binding sites for transcription factors that are downstream the pathways emanating from the BCR, including NF-κB signalling (NF-kB, BCL3 and OCT2), p38-JNK (JUNB, JUND, MEF2A, MEF2C and ATF2), RAS-RAF-MEK-ERK (CEBPB, SP1) and calcium-calmodulin (NFATC1). Chromatin regions that were more accessible in XPO1 mutated CLL were also enriched in binding sites for transcription factors that are regulated by the cytokine/inflammation signalling (STAT3 and STAT5) (Figure 1C). Chromatin regions that were less accessible in XPO1 mutated CLL were enriched in binding sites of FOXO1, whose nuclear localization is blocked by active BCR signalling.
The transcriptome of XPO1 mutated CLL cells is enriched in MAPK and inflammation signalling genes
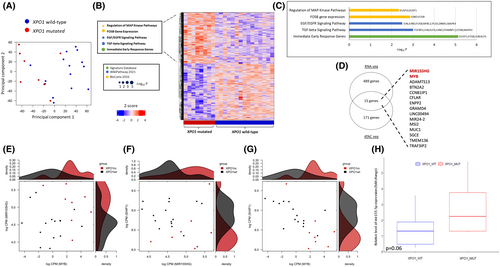
Overall, 236 genes were upregulated, and 296 genes were downregulated in XPO1 mutant CLL compared to XPO1 wild-type CLL. Consistently, by PCA, XPO1 mutated cases showed a distinct transcriptomic profile compared to XPO1 wild-type cases (p = 0.0126; Figure 2A,B). In keeping with the chromatin accessibility changes observed by ATAC-seq, XPO1 mutated CLL cells showed several transcriptomic features associated with BCR and cytokine signalling. Among pathways enriched in the upregulated genes, these included immediate early response to BCR activation, TGFβ signalling, EGF-EGFR signalling, FOSB gene expression and regulation of MAPK through DUSP (Figure 2C). The DUSP1 nuclear phosphatase, that inhibits JUN/FOS signalling, was upregulated in XPO1 mutant CLL and represents a potential candidate as cargo protein for XPO1. Using the Wregex prediction tool,26 we identified two different NES in DUSP1. Both NES show a negative charge in the amino-acid C-terminal residues (Table S4), suggesting that DUSP1 could better interact with XPO1E571K mutations.
The ATAC-seq and RNA-seq profiles of XPO1 mutant CLL identify upregulation of the miR-155/MYB pathway
By combining transcriptomic and epigenomic data, we identified 15 genes that are upregulated by RNA-seq and whose promoters are more accessible by ATAC-seq (Figure 2D). These genes include MIR155HG, the host gene that generates miR-155, and MYB, a transcription factor that positively regulates miR-155 expression. Given their role in BCR signalling, MIR155HG and MYB were further investigated.27 As expected, higher levels of MIR155HG correlated with higher levels of MYB in XPO1 mutant CLL (p < 0.05; Figure 2E). miR-155 expression favours BCR signalling through several mechanisms, including the downmodulation of SHIP1,28 a phosphatase that inhibits the BCR cascade. Accordingly, an inverse correlation between MIR155HG and MYB levels with SHIP1 levels was observed in XPO1 mutant CLL (all p < 0.05; Figure 2F,G).
To confirm that a higher expression of MIR155HG correlates with higher miR-155 levels, we measured miR-155 by RT-qPCR in XPO1 mutated (n = 8) and wild type (n = 8) CLL matched for IGHV status and FISH karyotype. Quantitative analysis confirmed the higher expression of miR-155 in XPO1 mutated CLL compared to XPO1 wild-type CLL (p = 0.065; Figure 2H). In addition, the SPI1/PU.1 gene, a transcription factor that is inhibited by miR-155,29 was found to be significantly downregulated in XPO1 mutated cases (p = 0.0029), thus cross-validating the miR-155 overexpression in XPO1 mutated CLL.
Association between XPO1 mutations and TTFT
As XPO1 mutated CLL were marked by the BCR and proliferation programs, we asked if XPO1 mutated CLL patients are characterized by a more aggressive clinical course. A retrospective, training validation design was used for the analysis in three different independent cohorts. To purify the impact of XPO1 mutations on clinical proliferation of CLL, only early-stage patients initially managed with a watch-and-wait policy were considered. XPO1 was mutated in 8/276 (2.9%) patients of the 1st cohort, in 12/286 (4.2%) patients in the 2nd cohort and in 8/395 (2.0%) patients in the 3rd cohort. The complete clinical and biological profile of the three cohorts is reported in Table 1. As expected for patients with early-stage CLL, high-risk molecular features were rare.
| Characteristics | 1st cohort | 2nd cohort | 3rd cohort |
|---|---|---|---|
| Values | Values | Values | |
| Median age | 70.9 (62.1–76.9) | 67.0 (56.0–73.0) | >65 years 55.0% |
| Median lymphocytes/μL | 8250 | 4151 | NA |
| B2M mg/L | 2.1 (1.7–2.6) | NA | NA |
| Gender | |||
| Male | 143 (51.8%) | 125 (57.8%) | NA |
| Female | 133 (48.2%) | 91 (42.1%) | NA |
| 13q deletion | |||
| Yes | 141 (51.1%) | NA | NA |
| No | 131 (47.5%) | NA | NA |
| Trisomy 12 | |||
| Yes | 41 (14.9%) | 35 (12.2%) | 56 (14.2%) |
| No | 231 (83.7%) | 217 (75.9%) | 339 (85.8%) |
| 11q deletion | |||
| Yes | 14 (5.1%) | NA | 34 (8.6%) |
| No | 258 (93.5%) | NA | 361 (91.4%) |
| 17p deletion | |||
| Yes | 10 (3.6%) | 10 (3.5%) | 25 (6.3%) |
| No | 262 (94.9%) | 344 (85.3%) | 370 (93.7%) |
| IGHV mutational status | |||
| Mutated | 198 (71.7%) | 173 (60.5%) | 291 (74.0%) |
| Unmutated | 68 (24.6%) | 112 (39.2%) | 102 (26.0%) |
- Abbreviations: B2M, beta-2-microglobulin; IGHV, immunoglobulin heavy chain variable.
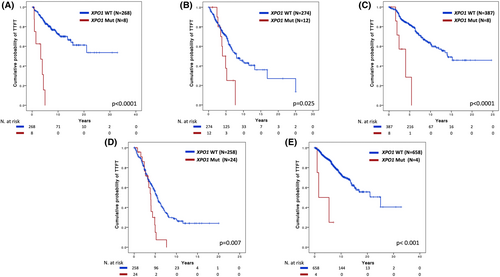
After a median follow-up of 10.4 years, XPO1 mutations were significantly associated with a shorter TTFT in the 1st cohort, with a TTFT at 10 years of 0% in XPO1 mutated patients compared to 69.8% in wild-type cases (Figure 3A) (HR 9.8, 95% CI 4.35–22.14, p < 0.001). In multivariate analysis including the IPS-E variables,18 which are validated for predicting TTFT and include unmutated IGHV genes, palpable lymph nodes and lymphocyte count >15 000/μL, XPO1 mutations maintained an independent association with a shorter TTFT (HR 2.79, 95% CI 1.16–6.71, p = 0.022; Table 2). XPO1 mutations maintained an independent association with a shorter TTFT (HR 3.56, 95% CI 1.48–8.59, p = 0.005) also after adjusting for variables of the Rai 0 prognostic score (Table 3)19 and NOTCH1 and SF3B1 mutations (Table S5). In the other two cohorts, XPO1 mutations were confirmed as a prognosticator of shorter TTFT. In the 2nd cohort, after a median follow-up of 5.6 years, the TTFT at 6 years was 25.0% in XPO1 mutated cases compared to 61.3% in XPO1 wild type cases (p = 0.025; HR 2.24, 95% CI 1.09–4.61, p = 0.029; Figure 3B). In the 3rd cohort, after a median follow-up of 6.7 years, the TTFT at 7 years was 0% in XPO1 mutated patients compared to 73.4% in wild-type patients (p < 0.0001; HR 6.02, 95% CI 2.48–15.03, p < 0.001; Figure 3C).
| Variable | HR | 95% CI | p value |
|---|---|---|---|
| Unmutated IGHV | 3.60 | 2.18–5.96 | <0.0001 |
| Palpable lymph nodes | 2.71 | 1.63–4.70 | <0.001 |
| Lymphocyte >15 000/μL | 2.49 | 1.50–4.11 | <0.001 |
| XPO1 mutations | 2.79 | 1.16–6.71 | 0.022 |
- Abbreviations: CI, confidence interval; HR, hazard ratio; IGHV, immunoglobulin heavy chain variable.
| Variable | HR | 95% CI | p value |
|---|---|---|---|
| WBC > 32 000/μL | 2.73 | 1.08–6.38 | 0.033 |
| Unmutated IGHV | 4.00 | 2.37–6.76 | <0.0001 |
| Del 17p | 1.63 | 0.49–5.42 | 0.493 |
| Tris 12 | 1.68 | 0.89–3.13 | 0.104 |
| Del 11q | 0.78 | 0.28–2.20 | 0.641 |
| XPO1 mutations | 3.56 | 1.48–8.59 | 0.005 |
- Abbreviations: CI, confidence interval; HR, hazard ratio; IGHV, immunoglobulin heavy chain variable; WBC, white blood cells.
By combining all cohorts (N = 957 patients), patients carrying either XPO1 E571 (N = 25) or D624 (N = 3) mutations showed superimposable outcomes in terms of TTFT (p = 0.594; Figure S3). In addition, the prognostic value of XPO1 mutations in terms of shorter TTFT was maintained in patients with both mutated and unmutated IGHV genes (Figure 3D,E).
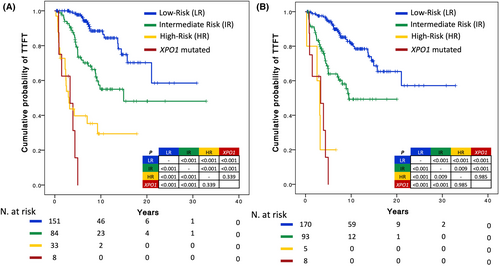
The IPS-E and Rai 0 models provide a benchmark of the expected TTFT in patients with high-risk early-stage CLL. We sought to understand whether early-stage CLL with XPO1 mutations has a similarly poor TTFT as early-stage CLL defined as high-risk by the IPS-E and Rai 0 models. The TTFT of early-stage patients harbouring XPO1 mutations, irrespective of their IPS-E score, did not significantly differ from the TTFT of early-stage patients belonging to the high-risk class according to IPS-E (Figure 4A). Similarly, the TTFT of early-stage patients harbouring XPO1 mutations, irrespective of their Rai 0 score, did not significantly differ from the TTFT of early-stage patients belonging to the high-risk class according to the Rai 0 model (Figure 4B). XPO1 mutations therefore identify patients with early treatment requirements independent of IPS-E and Rai 0 prognostic models.
DISCUSSION
Most CLL patients are diagnosed in an asymptomatic stage and are managed with a watch-and-wait strategy.10 However, their clinical outcome is heterogenous and different patients may progress and require therapy at different timepoints. In the present study using a training-validation approach, XPO1 mutations emerged as an independent predictor of shorter TTFT capturing a fraction of patients that require treatment soon after diagnosis. XPO1 mutant CLL cells are characterized by a higher expression of signalling pathways downstream to the BCR that may predispose to a higher proliferative behaviour.
To gain insights into the mechanisms that may drive the shorter TTFT and LDT of XPO1 mutated CLL, we evaluated the transcriptomic and the epigenomic profile of XPO1 mutated CLL and compared it to that of wild type cases matched for IGHV status, FISH karyotype and TP53 status. BCR signalling appeared as a potential candidate that might contribute, at least in part, to the higher proliferation rate of XPO1-mutated CLL. In fact, chromatin regions that were more accessible in XPO1 mutated CLL were enriched in binding sites for transcription factors that act downstream to pathways emanating from the BCR. These pathways include NF-κB signalling, p38-JNK, RAS–RAF–MEK–ERK and calcium-calmodulin. In addition, the combination of RNAseq and ATAC-seq data identified the overexpression and the increased promoter accessibility of the MYB and MIR115HG genes in XPO1 mutated CLL compared to wild-type cases. Importantly, MYB is known to induce expression of MIR115HG, the gene coding for miR-155 that inhibits the expression of SHIP-1 and, consequently, contributes to stimulate the activity of the BCR. These results provide novel evidence in primary CLL cells on the association between XPO1 mutation and an enrichment in pathways regulating lymphocyte activation, NFAT signalling and NF-κB activation. Also, these data integrate and expand previous suggestions on the association of XPO1 mutations with genes of the inflammation and cytokine signalling pathways.5, 30
The precise mechanisms by which XPO1 mutations induce the upregulation of miR-155 and of other signalling pathways remain currently unknown. In our study, DUSP1, encoding a nuclear phosphatase that inhibits JUN/FOS signalling, has been identified by RNA-seq as one of the major upregulated genes in XPO1 mutant CLL patients. The Wregex prediction model generated in our study has identified two different negatively charged NES domains in the DUSP1 protein, rendering it a candidate of increased export out of the nucleus in the presence of XPO1 mutations that enhance export of proteins bearing a negative NES domain. Nuclear export of DUSP1 may in turn favour JUN/FOS signalling in the nucleus. Since the MIR155HG promoter contains a conserved binding site for JUN/FOS,31 an enhanced signalling of this pathway could promote the transcription of MIR155HG, that would promote BCR signalling through miR-155. In vitro, studies will be required to confirm the hypothesis generated by our current data.
Since the transcriptomic and epigenomic profile of XPO1 mutated CLL pointed to pathways that promote BCR signalling and may affect CLL proliferation, we analysed the correlation of XPO1 mutations with TTFT. Previous reports demonstrated conflicting results regarding the prognostic impact of XPO1 mutations as a marker for shorter TTFT conceivably because they included CLL patients at different clinical stages.20, 30, 32 In this study, by including only early-stage CLL (Rai 0/I and Binet A), XPO1 mutations were sorted out as a validated prognosticator of shorter TTFT. Importantly, the prognostic role of XPO1 mutations in anticipating TTFT in early-stage CLL is independent of the IGHV status and of the variables included in the two prognostic scores for early-stage CLL patients.18, 19 Since two unique codons are the hotspots affected by XPO1 mutations in CLL, primer-based methods may be used to identify XPO1 mutations in a simple and time effective manner. Prediction of TTFT may be important to identify high-risk patients that might be suitable for early intervention clinical trials and to reassure patients with a very indolent disease that may be followed with a less intense schedule.18, 19
Finally, the longitudinal analysis of CLL samples harbouring XPO1 mutations showed that the XPO1 mutant clone did not undergo clonal expansion either during watch & wait management or after therapy, including chemoimmunotherapy and ibrutinib. This observation may suggest that XPO1 mutations are not selected by treatment. Conversely, the acquisition of XPO1 mutations was observed in the lymph node biopsy of patients who transformed to clonally related Richter syndrome, suggesting that XPO1 mutations might represent a novel mechanism involved in Richter transformation.
AUTHOR CONTRIBUTIONS
Gianluca Gaidano, Davide Rossi and Riccardo Moia designed the study, interpreted data, and wrote the manuscript; Valter Gattei and Robin Foà contributed to study design and manuscript revision; Lodovico di Terzi Bergamo, Donatella Talotta, Riccardo Bomben, Gabriela Forestieri, Valeria Spina, Alessio Bruscaggin, Tamara Bittolo and Antonella Zucchetto performed molecular studies and contributed to manuscript revision; Chiara Cosentino, Mohammad Almasri, Riccardo Dondolin, Silvia Rasi, Abdurraouf Mahmoud, Wael Al Essa, Bassel Awikeh, Sreekar Kogila, Matteo Bellia and Samir Mouhssine contributed to data analysis and manuscript preparation; Stefano Baldoni, Ilaria Del Giudice, Francesca Romana Mauro, Rossana Maffei, Annalisa Chiarenza, Agostino Tafuri, Roberta Laureana, Maria Ilaria Del Principe, Francesco Zaja, Giovanni D'Arena, Jacopo Olivieri, Paolo Sportoletti, Roberto Marasca, Lydia Scarfò and Paolo Ghia provided study material and contributed to manuscript revision.
ACKNOWLEDGEMENTS
This work was supported by: Molecular bases of disease dissemination in lymphoid malignancies to optimize curative therapeutic strategies, (5 × 1000 No. 21198), Associazione Italiana per la Ricerca sul Cancro Foundation Milan, Italy; Progetti di Rilevante Interesse Nazionale (PRIN; 2015ZMRFEA), Rome, Italy; the AGING Project—Department of Excellence—DIMET, Università del Piemonte Orientale, Novara, Italy; and Ricerca Finalizzata 2018 (project RF-2018-12365790), MoH, Rome, Italy; Swiss Cancer League, ID 3746, 4395 4660, and 4705, Bern, Switzerland; Research Advisory Board of the Ente Ospedaliero Cantonale, ABREOC 2019-22514, Bellinzona, Switzerland; European Research Council (ERC) Consolidator Grant CLLCLONE, ID: 772051; Swiss National Science Foundation, ID 320030_169670/1 and 310030_192439, Berne, Switzerland; Fondazione Fidinam, Lugano, Switzerland; Nelia & Amadeo Barletta Foundation, Lausanne, Switzerland; Fond'Action, Lausanne, Switzerland; The Leukemia & Lymphoma Society, Translational Research Program, ID 6594-20, New York. The authors are grateful to Ahad Ahmed Kodipad and Chiara Favini for their technical help with this study.
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interests regarding this manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.