Case report: Rare case of donor cell-derived T-cell acute lymphoblastic leukaemia in a female patient after receiving an allo-transplant from her male sibling
Summary
Donor-derived haematological neoplasms, in which recipients present with haematological malignancies that have evolved from transplant donor stem cells, have previously been described for myelodysplastic syndrome, myeloproliferative neoplasms, acute myeloid leukaemia and less often, leukaemias of lymphoid origin. Here we describe a rare and complex case of donor-derived T-cell acute lymphoblastic leukaemia with a relatively short disease latency of less than 4 years. Through genomic and in vitro analyses, we identified novel mutations in NOTCH1 as well as a novel activating mutation in STAT5B; the latter targetable with the clinically available drugs, venetoclax and ruxolitinib.
INTRODUCTION
Clonal haematopoiesis (CH) is the expansion of haematopoietic stem cells (HSCs) harbouring somatic mutations associated with blood cancers and occurs more frequently in people aged over 60.1 However, it leads to the development of haematological neoplasms including myelodysplastic syndrome (MDS), myeloproliferative neoplasms and acute myeloid leukaemia (AML). The majority of people exhibiting CH never progress to overt disease. There is some evidence for a familial predisposition in CH2; however, two recent twin studies suggested additional important contributions from non-genetic factors.3, 4
DTA mutations, loss-of-function mutations in the epigenetic modifiers DNMT3A, TET2, and ASXL1, are frequently observed in CH. Large cohort studies have demonstrated HSCs harbouring DNMT3A mutations precede the onset of AML and MDS.5 Furthermore, those who do develop myeloid malignancies often have germline mutations in leukaemic driver genes.6 Donor-engrafted CH following stem cell transplant (SCT) occurs less frequently. The occurrence of a donor cell-derived haematological neoplasm (DCHN) is even rarer, with approximately 150 cases of DCHN reported thus far, the majority with limited genomic data.7
Here, we report a rare and complex case of donor cell-derived T-cell acute lymphoblastic leukaemia (T-ALL), following sex-mismatched SCT, in a female patient with a history of haematological malignancies. Transcriptomic analyses revealed two novel NOTCH1 frameshift mutations, and a novel STAT5B single nucleotide variant (SNV) that potentially contributed to T-ALL relapse in this patient. Furthermore, ex vivo experiments demonstrated that cells harbouring the STAT5B mutation are sensitive to the JAK inhibitor ruxolitinib and the Bcl-2 inhibitor venetoclax, highlighting the benefit of a precision medicine approach in the treatment of relapsed T-ALL with constitutively active JAK–STAT signalling.
CASE REPORT
In 2011, a 38-year-old female patient presented with stage IVA Diffuse Large B-cell Lymphoma (DLBCL) involving the bone marrow, lymph nodes and right breast. She achieved remission after six cycles of immunochemotherapy (R-CHOP: rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) with high-dose methotrexate as central nervous system (CNS) prophylaxis. In 2013, she developed treatment associated AML (t-AML) accompanied by t(8;21). The patient achieved remission with 7 + 3 (cytarabine/idarubicin) induction chemotherapy. This was followed by high-dose cytarabine consolidation prior to SCT in 2014 using peripheral blood stem cells (PBSC) from her 42-year-old brother with myeloablative (busulfan/cyclophosphamide) conditioning. Complete donor chimerism was achieved 1 month after transplant (Figure S1). In 2018 the patient, then 45 years, presented with cervical lymphadenopathy, leukaemic infiltrates in the lymph nodes and bone marrow, and was diagnosed with T-ALL (T-ALL presentation sample). Cytogenetic, genomic (Multiplex Ligation-dependent Probe Amplification [MLPA]) and transcriptomic analyses (mRNA-Seq) identified that the leukaemic cells were of male origin, confirming that the T-ALL was donor cell-derived (Figures S2 and S3). The patient achieved a morphological complete remission after hyper-CVAD combination chemotherapy but relapsed 4 months later (T-ALL relapse sample). The patient had further salvage with FLAG chemotherapy (fludarabine, cytarabine, G-CSF) before achieving morphological remission with nelarabine. Intrathecal chemotherapy was given with each cycle as CNS prophylaxis. She proceeded to matched unrelated allogeneic SCT using PBSC after reduced-intensity conditioning with fludarabine, melphalan and thymoglobulin. Four months post-second transplant, the patient developed vision loss and progressive facial nerve palsy. Magnetic resonance imaging and cerebrospinal fluid examination confirmed a second relapse in the CNS. The patient received intrathecal chemotherapy for palliation and died of relapsed disease aged 46 years, 14 months after the initial T-ALL presentation. The sibling donor is currently well.
RESULTS AND DISCUSSION
Cytogenetic analysis of leukaemic blasts at T-ALL presentation showed a male karyotype, with a pathogenic clone characterised by partial deletion of chromosome 9p13 and a t(11;14)(p12–14;q32) translocation. Additional karyotypic details are available in Supplementary Note 1.
Transcriptomic analyses were performed on bone marrow samples collected at the time of T-ALL presentation, as well as first relapse, and analysed using our bioinformatic pipelines for the detection of gene fusions, SNVs, insertion/deletion alterations (INDELs) and assessment of gene expression.8 mRNA-Seq identified several non-synonymous SNVs and INDELs of clinical relevance (Table S1; Figure 1A), which were validated by PCR and Sanger sequencing (Figure S4). The KMT2D p.L2610P mutation was observed at both T-ALL presentation (variant allele frequency [VAF] = 0.55) and relapse (VAF = 0.60). Investigation of sibling donor material and patient mesenchymal stem cells (MSCs) to determine germline mutation status revealed KMT2D p.L2610P was present in both, indicating this mutation is likely familial (Figure S4). A germline mutation screening panel identified a single germline silent mutation in the XPC gene and no somatic mutations in any of the other 67 included genes (Table S2). KMT2D is an epigenetic modifier that regulates the expression of genes involved in development, differentiation and tumour suppression. While the specific function of the p.L2610P mutation is unknown, KMT2D mutations have been widely reported in cancers, including haematological malignancies, and likely play an important role in cancer initiation and/or progression.
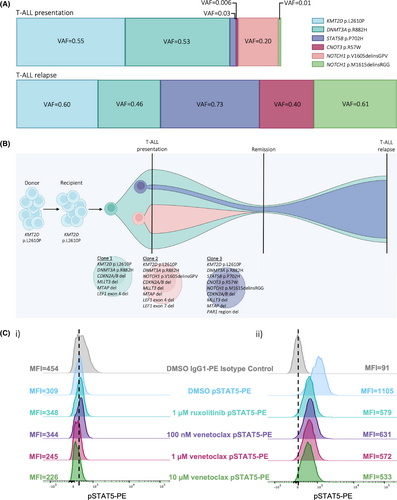
The DNMT3A p.R882H mutation, identified at similar VAF at both presentation and relapse, was absent in donor material (Figure S4). Mutations in this epigenetic regulator are a hallmark of age-associated CH. The p.R882H dominant negative mutation disrupts DNMT3A function and is rarely observed in patients <49 years old,2, 5 as in our case where the patient was 45 years at T-ALL presentation. Similar to other reports, we used the VAF of somatic mutations in sequential samples to study leukaemic clonal evolution9, 10 using the fishplot package for R.11 It is likely that DNMT3A p.R882H was acquired in a clone harbouring KMT2D p.L2610P shortly after transplant (Clone 1). These mutations, combined with the increased proliferative and self-renewal stress associated with the engraftment process, presumably facilitated the acquisition of mutations in NOTCH1, STAT5B and CNOT3 and the subsequent development of T-ALL (Clones 2 and 3). The potential evolution of mutation acquisition is summarised in Figure 1B. The NOTCH1, STAT5B and CNOT3 mutations were not identified in donor material or the patient's MSCs (Figure S4).
Two novel NOTCH1 INDEL mutations were identified on separate alleles (i.e., in separate leukaemic clones) in a previously defined heterodimerisation domain mutation hotspot (Figures S4 and S5). NOTCH1 p.V1605delinsGPV (VAF = 0.20) was identified in the presentation sample only (Clone 2). Interestingly, a second NOTCH1 INDEL mutation, p.M1615delinsRGG, was identified at low frequency at presentation (VAF = 0.01, Clone 3), then expanded at relapse (VAF = 0.61). There was no evidence of p.V1605delinsGPV in the relapse sample, suggesting expansion of the minor leukaemic Clone 3 as the patient's disease progressed. Both NOTCH1 mutations identified are likely to be activating mutations leading to constitutive Notch signalling.12
The STAT5B p.P702H mutation was observed at low frequency at T-ALL presentation (VAF = 0.03) followed by expansion at relapse (VAF = 0.73), suggesting it resides in Clone 3. The p.P702 residue lies in the SH2 domain responsible for the recruitment of STAT5 molecules to activated cytokine receptors (JAKs) with subsequent phosphorylation and dimerisation.13 While this mutation has not been previously reported, perturbation of this residue likely results in increased activation of STAT5 (pSTAT5). Indeed, a marked increase in pSTAT5 in bone marrow mononuclear cells (BMMNCs) isolated from the relapse sample was observed compared with pSTAT5 in the presentation BMMNCs (mean fluorescence intensity [MFI] = 1105 vs. 309, respectively; Figure 1C). A 2-h incubation of BMMNCs with the Jak1/2 inhibitor ruxolitinib led to normalisation of pSTAT5 (MFI = 1105 to 579). Leukaemic BMMNCs also demonstrated dose-dependent sensitivity to venetoclax comparable to that observed for ruxolitinib (Supplementary Note 2; Figure 1C).
Mutations in the tumour suppressor CNOT3, including the p.R57W alteration identified here, have previously been reported in T-ALL.12 As with the NOTCH1 p.M1615delinsRGG and STAT5B p.P702H variants, CNOT3 p.R57W was observed at very low frequency at presentation, then expanded at relapse (VAF = 0.006 vs. 0.40), suggesting a close temporal acquisition of all three mutations in Clone 3. Given that constitutive activation of Notch signalling alone is unable to induce self-renewal of T-cell progenitors,14 we speculate that the activating STAT5B mutation likely increased pathogenicity through JAK/STAT signalling, though we were unable to study this further with techniques such as single-cell sequencing due to limited samples.
DNA copy number variations (CNVs) were detected via MLPA (MRC Holland) (Figure S2). Results demonstrated deletions in the CDKN2A/B, MLLT3, MTAP and LEF1 genes (further discussed in Supplementary Note 3).
CONCLUSION
Here we describe a patient with a history of haematological malignancies (DLBCL and t-AML) who developed T-ALL following allogeneic SCT. We identified two novel NOTCH1 INDEL mutations and a novel STAT5B mutation. Donor cell-derived leukaemia is a rare complication of SCT, usually resulting in leukaemias of myeloid origin, with an estimated incidence of 0.1%.15 While the aetiology of donor cell-derived leukaemias is unclear, evidence suggests the transfer of pre-leukaemic clones from donor to recipient,16 leading to stochastic acquisition of additional alterations and disease onset in susceptible recipients. As in the current case, the majority of the time, the donor does not go on to develop leukaemia, suggesting that it is the recipient's microenvironment providing favourable conditions for leukaemogenesis. Indeed, the donor was relatively young (42 years old) and at low risk of CH.5 Combined with the patient's history of two additional haematological neoplasms, the disease trajectory suggests her bone marrow microenvironment was pre-disposing for leukaemic transformation, in keeping with the ‘two-hit’ model of acute lymphoblastic leukaemia development.17 While no material is available to examine the bone marrow niche in more detail, we hypothesise that KMT2D p.L2610P was the ‘first-hit’ pre-transplant. Combined with chemotherapy and transplant-related cellular stress, this created a microenvironment amenable to acquiring the ‘second-hit’, the DNMT3A p.R882H mutation. Mutations in these two epigenetic regulators presumably resulted in (1) alteration of expression of genes involved in development, differentiation and tumour suppression (2) selection of a clone harbouring alterations favourable for T-ALL pathogenesis (CDKN2A/B, MLLT3 and LEF1 deletions). Combined, these factors lead to the acquisition of mutations in the T-ALL-associated genes NOTCH1, STAT5B and CNOT3 and the onset of an aggressive disease and subsequent relapse within 14 months.
AUTHOR CONTRIBUTIONS
Laura N. Eadie designed and performed experiments, analysed data, interrogated and evaluated the bioinformatic output, wrote the manuscript, created the figures. Jacqueline A. Rehn performed bioinformatic analyses of transcriptomic sequencing and created the figures. Caitlin E. Schutz designed and performed experiments, analysed data. Susan L. Heatley designed and performed experiments and evaluated the bioinformatic output. Monika M. Kutyna designed and performed experiments. Devendra K. Hiwase provided clinical data and interpretation. Deborah L. White provided oversight of experimental and bioinformatic design, evaluated the bioinformatic output and wrote the manuscript. David T. Yeung provided clinical data and interpretation, evaluated the bioinformatic output and wrote the manuscript. All authors critically revised and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
Thank you to Phuong Dang for assistance with PCR primer design and provision of nested PCR protocol. The authors would like to thank Genetics and Molecular Pathology at SA Pathology, Adelaide, SA, Australia for performing cytogenetic and FISH analyses. Open access publishing facilitated by The University of Adelaide, as part of the Wiley - The University of Adelaide agreement via the Council of Australian University Librarians.
FUNDING INFORMATION
This work was supported by the Peter Nelson Leukaemia Research Fellowship Fund administered by the Cancer Council South Australia (022212 to LNE). LNE is the Peter Nelson Leukaemia Research Fellow. SLH is a The Kids' Cancer Project Fellow. DKH is a MRFF Investigator Grant recipient. DLW is a National Health and Medical Research Council (NHMRC) Career Development Fellow and a Cancer Council South Australia Beat Cancer Project Principal Research Fellow. DTY is a NHMRC Early Career Fellow. Funding from NHMRC, MRFF, Bristol-Myers Squibb (to DLW).
CONFLICT OF INTEREST STATEMENT
LNE, CES, JAR, SLH and MMK have no conflicts of interest to declare. DKH is on advisory committees for Novartis and AbbVie. DLW receives honoraria and research funding from Bristol-Myers Squibb and honoraria from Amgen. DTY receives honoraria and research funding from Bristol-Myers Squibb and Novartis and honoraria from Amgen and Pfizer.
ETHICS STATEMENT
This study was approved by the Women's and Children's Health Network Human Research Ethics Committee (2023/HRE00071). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
PATIENT CONSENT STATEMENT
Informed consent was obtained from the patient included in the study.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. For original data, contact [email protected].




