A three-miRNA-based expression signature at diagnosis can predict occurrence of relapse in children with t(8;21) RUNX1-RUNX1T1 acute myeloid leukaemia
Children with acute myeloid leukaemia (AML) harbouring CBF anomalies notoriously have a more favourable prognosis than other AML cytogenetic subgroups. In the Associazione Italiana di Ematologia e Oncologia Pediatrica (AIEOP)-AML 2002/01 protocol, these patients were allocated to the standard-risk (SR) group and all achieved morphological complete remission (CR) after induction therapy (Pession et al, 2013; Manara et al, 2014). However, approximately one-third of them – in particular those harbouring t(8;21)RUNX1/RUNX1T1 – relapsed (Pession et al, 2013). These results prompted us to refine the definition of SR, trying to identify those t(8;21) AML patients who could be pre-emptively directed to an increased chemotherapy treatment or to allogeneic haematopoietic stem cell transplantation to properly tackle their higher risk to relapse. To this purpose, we performed, for the first time, a retrospective analysis to identify a prognostic model, based on micro RNA (miRNA) expression, capable of enhancing the prediction of recurrence in paediatric t(8;21) AML patients. We determined the miRNA expression profile of 33 patients enrolled in AIEOP AML 2002/01. In our cohort, 14 out of 33 (42·4%) patients suffered relapse after a median time of 280 days and 6 of them died, while only 2 out of the 19 non-relapsed patients died (42·8% vs. 10·5%; Fisher test P < 0·05). We identified 115 differentially expressed (DE) miRNAs between relapsed and non-relapsed patients and the miRNA profile was able to dichotomize patients according to their relapse status (Fig 1A, Kruskal–Wallis test, P < 0·05; Table SI). We divided the entire cohort of patients in two subgroups: one was defined as the training set (n = 17), and the remaining 16 patients constituted the validation set. The two sets were similar for main clinical and biological parameters (Table SII). We used a univariate model (see Supplementary Methods) where each miRNA of the microarray platform was studied for its association with the event relapse (CIR, Cumulative Incidence of Relapse; Gray Test P < 0·05) in both training and validation sets (Fig 1B). We independently confirmed these results through a multivariate Cox proportional hazards model that included the two other variables impacting on CIR: KIT mutational status and age at diagnosis (Table SIII, P = 0·098 and P = 0·096; Fig S1), and by ROC analysis (Fig S2).
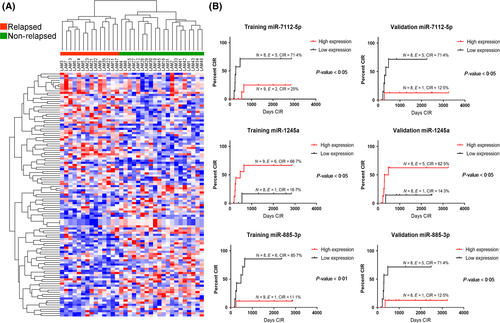
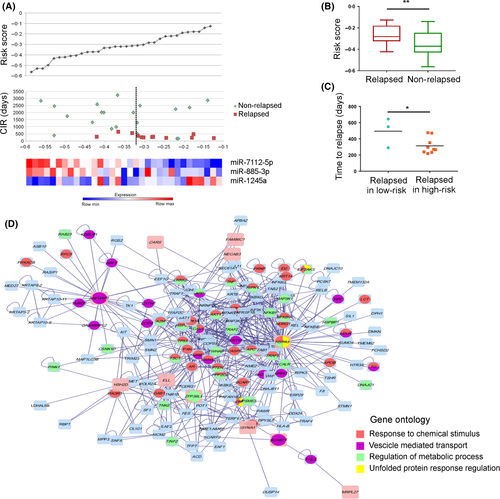
We identified a three-miRNA signature, involving miR-7112-5p, miR-885-3p and miR-1245a, able to predict already at diagnosis a different risk of disease recurrence among t(8;21)RUNX1/RUNX1T1 patients. Then, by using miRNA expression levels and the CIR data we obtained a patient miRNA-based risk score (Fig 2A), which showed that relapsed patients had a significantly higher risk score as compared to non-relapsed patients (unpaired t-test, P < 0·01; Fig 2B). Moreover, we observed that relapsed patients with a high-risk score suffered relapse earlier after diagnosis than patients who had a lower risk score (313 days vs. 495 days, t-test P < 0·05; Fig 2C). The expression of these three-miRNAs did not change in 6 patients' matched specimens collected at both diagnosis and relapse (paired t-test; Fig S3), suggesting that they may characterize the smaller subclone which escapes chemotherapy and is responsible for disease relapse. To obtain more information on the pathways controlled by these three-miRNAs, we performed gene expression profiles in 16 out of 33 t(8;21) patients of the same cohort (Tregnago et al, 2016) (GSE75461), identifying 281 DE genes (multiple t-test, P < 0·01; Table SIV) able to separate patients in two groups according to the three-miRNA risk score: 9 patients at high-risk and 7 patients at low-risk (Fig S4). We performed gene functional enrichment using the Reactome Pathways Database (http://reactome.org/) and determined that the most deregulated genes were part of the metallothioneins, cell metabolism and TP53 pathways (Hyper-geometric test, P < 0·05; Table SV), which may enable specific malignant subclones to gain a proliferative advantage during treatment, mostly by decreasing sensitivity to chemotherapy. Then, considering the two array platforms, we performed gene expression profiling to predict miRNA targets by the DIANA-mirPath tool (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index). Three hundred and twenty-two genes were putative targets of the three-miRNAs, 17 of which were significantly associated with the risk of recurrence at the same level of miRNA expression. Finally, we confirmed 14 putative pairs of miRNA and mRNA targets with opposite correlated expression predicting the risk of relapse (Table SVI). In particular, one pair showed down-regulation of MRPL27 and upregulation of miR-1245a; the remaining pairs showed upregulation of the gene correlating with the downregulation of the miRNA (Table SVI). These data were used in five different protein–protein interaction databases; we found that this three-miRNA signature was informative of the t(8;21)-rearranged AML with more aggressive features. The schema (Fig 2D) visualized the tangled network of the aberrant transcriptional circuit related mostly to cellular stress and unfolded protein trafficking that occurred in t(8;21) patients who relapsed. For instance, this novel three-miRNA signature takes in account miR-885-3p, previously identified to be an onco-suppressor in colon cancer xenografts and to regulate apoptosis and autophagy after cisplatin exposure (Huang et al, 2011; Xiao et al, 2015). High miR-1245a expression has been found in exosomes isolated from plasma samples of AML/myelodysplasia patients that influence immune-surveillance, as well as in breast cancer cells targeting BRCA2 and impairing DNA repair (Song et al, 2012). Herein, the most connected gene in the network, HSPA5 is predicted to be a target of two out of our three miRNAs, namely miR-885-3p and miR-7112-5p. HSPA5 is a member of the Heat shock protein 70 protein family, which protects cancer cells against the adverse hypoxic and nutrient-deprived microenvironment in solid tumours (Lee, 2007). Interaction with miR-885-3p has been previously validated in the bone marrow microenvironment (Balakrishnan et al, 2014) and the overexpression of HSPA5 mediated resistance to cytarabine-induced apoptosis, supporting a central role in the chemo-resistance of t(8;21) patients to be further considered (Uckun et al, 2011). In summary, the present study represents the most numerous paediatric t(8;21) AML miRNA profiles and identifies a unique three-miRNA signature relevant for paediatric t(8;21) AML patients that, when present at diagnosis, can predict a higher risk of relapse. miRNA expression showed a remarkable prognostic performance with respect to other clinical and biological factors, supporting further in vitro studies to pursue new tailored therapeutic approaches in t(8;21) AML patients at higher risk of recurrence.
Acknowledgements
We thank the members of all of the AIEOP centres, and Katia Polato and AnnaMaria Di Meglio, of the Haematology-Oncology Clinic at UniPD, for their work on identification of chromosomal aberrations in children treated in AIEOP centres.
Authorship contributions
MZ was involved in the concept, design, data analysis and interpretation of this study and wrote the manuscript. VB performed array experiments; AL performed karyotypes for AEIOP-AML; MCP, CR, GM collected samples in AIEOP centres. AP was the principal investigator of the concluded AML 2002/01 protocol. FL is the principal investigator of the AML 2013/01 protocol and chairperson of the AIEOP-AML working group. FL, MP, GB, CT were involved in design and data interpretation, writing and critical evaluation of this manuscript.
Conflict of interest disclosure
The authors declare no competing financial interests.
Funding sources
This work was supported by grants from CARIPARO Istituto di Ricerca Pediatrica-Fondazione Città della Speranza and Università degli Studi di Padova and by AIRC (Associazione Italiana Ricerca sul Cancro), special grant 5x1.000.