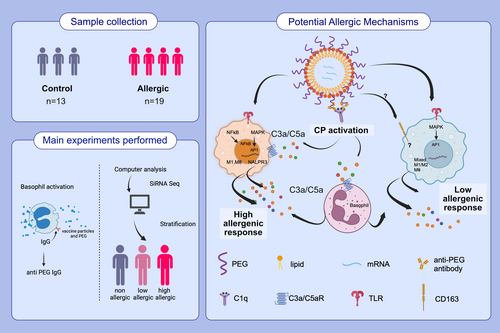
Elucidating allergic reaction mechanisms in response to SARS-CoV-2 mRNA vaccination in adults
Mihir M. Shah, Janice A. Layhadi, and Dennis E. Hourcade contributed equally to this work.
Mohamed H. Shamji, Christine T.N. Pham, and Kari C. Nadeau contributed equally to this work.
Abstract
Background
During the COVID-19 pandemic, novel nanoparticle-based mRNA vaccines were developed. A small number of individuals developed allergic reactions to these vaccines although the mechanisms remain undefined.
Methods
To understand COVID-19 vaccine-mediated allergic reactions, we enrolled 19 participants who developed allergic events within 2 h of vaccination and 13 controls, nonreactors. Using standard hemolysis assays, we demonstrated that sera from allergic participants induced stronger complement activation compared to nonallergic subjects following ex vivo vaccine exposure.
Results
Vaccine-mediated complement activation correlated with anti-polyethelyne glycol (PEG) IgG (but not IgM) levels while anti-PEG IgE was undetectable in all subjects. Depletion of total IgG suppressed complement activation in select individuals. To investigate the effects of vaccine excipients on basophil function, we employed a validated indirect basophil activation test that stratified the allergic populations into high and low responders. Complement C3a and C5a receptor blockade in this system suppressed basophil response, providing strong evidence for complement involvement in vaccine-mediated basophil activation. Single-cell multiome analysis revealed differential expression of genes encoding the cytokine response and Toll-like receptor (TLR) pathways within the monocyte compartment. Differential chromatin accessibility for IL-13 and IL-1B genes was found in allergic and nonallergic participants, suggesting that in vivo, epigenetic modulation of mononuclear phagocyte immunophenotypes determines their subsequent functional responsiveness, contributing to the overall physiologic manifestation of vaccine reactions.
Conclusion
These findings provide insights into the mechanisms underlying allergic reactions to COVID-19 mRNA vaccines, which may be used for future vaccine strategies in individuals with prior history of allergies or reactions and reduce vaccine hesitancy.
Graphical Abstract
We analyze some of the mechanisms underlying allergic reactions to COVID-19 mRNA vaccines. After performing hemolysis, indirect basophil activation, and single-cell multiome assays, we elucidated certain allergic reaction mechanisms. We found anti-PEG IgG correlates with vaccine-mediated complement activation and that epigenetic modulation of mononuclear phagocytes could influence vaccine reaction manifestations.Abbreviations: AP-1, activator protein 1; C1q, complement component 1q; C3, complement component 3; COVID-19, coronavirus disease 2019; CP, classical pathway; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor kappa-light-chain-enhancer; NLRP3, nucleotide-binding domain, leucine-rich–containing family, pyrin domain–containing-3; PEG, polyethylene glycol; siRNA seq, small interfering RNA sequencing; TLR, Toll-like receptor.
Abbreviations
-
- AP-1
-
- activator protein 1
-
- C1q
-
- complement component 1q
-
- C3
-
- complement component 3
-
- COVID-19
-
- coronavirus disease 2019
-
- CP
-
- classical pathway
-
- MAPK
-
- mitogen-activated protein kinase
-
- NF-κB
-
- nuclear factor kappa-light-chain-enhancer
-
- NLRP3
-
- nucleotide-binding domain, leucine-rich-containing family, pyrin domain-containing-3
-
- PEG
-
- polyethylene glycol
-
- siRNA seq
-
- small interfering RNA sequencing
-
- TLR
-
- Toll-like receptor
1 INTRODUCTION
The mRNA vaccines, BNT162b2 (Pfizer-BioNTech) and mRNA-1273 (Moderna), have been proven to be effective and safe, with approximately 95% efficacy in preventing COVID-19 in early studies against the original Sars-CoV-2 strains.1-4 However, a small portion of the population has experienced allergic reactions resulting from vaccination, specifically 11.1 cases of anaphylaxis per million doses for the BNT162b2 vaccine and 2.5 cases of anaphylaxis per million doses for the mRNA-1273 vaccine.1-4 Despite these findings, the mechanisms underlying these allergic reactions have yet to be characterized.
The COVID-19 mRNA vaccines each utilize different liposomal delivery vehicles that contain PEG2000.5 Polyethylene glycol (PEG) is used in many drug and vaccine formulations in addition to cosmetics and lotions, to improve water solubility. Reports have shown that up to 70% of patients who received PEGylated formulations develop anti-PEG antibodies.6 It is hypothesized that allergic reactions to COVID-19 vaccines could be due to a pre-existing anti-PEG allergy,5, 7 although true IgE-mediated reactions to PEG in COVID-19 mRNA vaccines have not been definitively proven.
The complement system presents a rapid-acting, first-line defense of the intravascular space and other biological compartments from foreign invaders and facilitates the safe removal of apoptotic cells, immune complexes, and cellular debris.8 Early evidence suggesting that nanoparticles can activate complement came from the clinical use of Doxil®, a PEGylated liposomal formulation of doxorubicin for cancer treatment.9, 10 Patients typically exhibited signs of cardiopulmonary distress that developed immediately after the start of infusion, including dyspnea, tachypnea, tachycardia, hypotension, and chest and back pain. These symptoms coincided with a rise in the plasma concentration of complement activation products and typically occurred within minutes upon first exposure to the drug.11 Subsequent animal studies demonstrated that reaction to Doxil® was triggered by anti-PEG IgM-induced complement activation, and this phenomenon became known as complement activation-related pseudoallergy or CARPA.9, 10, 12 Since that time, the spectrum of reagents that cause pseudoallergic symptoms has been broadened to include micelle-solubilized drugs, certain antibodies, and contrast media, in addition to PEGylated formulations and other liposomal drugs.13
Additionally, most studies on COVID-19 mRNA allergic reactions have focused mainly on the types of immune responses while genetic predisposition is so far limited to testing for alpha tryptasemia.14 For those that consented and were tested, no genetic predispositions to alpha-tryptasemia were found, prompting us to further examine the genetic makeup of allergic individuals. Allergies and allergic reactions are complex processes involving both genetics and the environment factors that affect gene expression without altering the DNA sequence, resulting in epigenetic traits that modulate the immune response plasticity.15-17 In this study, we explored the role of epigenetics in COVID-19 mRNA reactions as contributing factors to the variable vaccine reaction predisposition.
2 METHODS
2.1 Study Design
From December 2020 to April 2021, individuals who called the Stanford University or Washington University allergy clinics to consult for possible allergic reactions to the COVID-19 mRNA vaccines and who met the definition of immediate (within 2 h of vaccine receipt) allergic reaction as per our prior publication18 were invited to participate in our IRB-approved studies. Thirty-two participants (19 allergics and 13 controls) were enrolled for this study under IRBs approved by Stanford University Institutional Review Board (IRB 8269) and the Washington University in St. Louis Institutional Review Board (IRB 202101196). They were consented at different time points (0–86 days) after their allergic reactions and asked to provide up to 3 blood samples on separate days after their first dose of vaccination. Due to the nature of the COVID-19 pandemic at the time, there was a necessity to immediately vaccinate individuals; thus, it was not possible to collect blood samples prior to vaccination and allergic reaction. Individuals <18 yo were not included since the vaccines were not approved for that age group at the time of study. None of the individuals had tryptase levels and/or CH50 levels drawn. These blood samples were utilized to perform all the experiments outlined within the study. The experiments were performed without randomization or blinding. 10 out of 19 allergic reaction participants were tested and were negative for an alpha tryptasemia mutation to ensure allergic symptoms post vaccination were not caused by an elevated copy number of TPSAB1 encoding alpha tryptase.14 Individuals within the nonallergic group have ID numbers of N-X, where X is a number between 1 and 13, and individuals within the allergic group have ID numbers of A-X, where X is a number between 1 and 19. This entire study was conducted within full compliance of good clinical practice.
2.2 Anti-PEG-IgG and IgE ELISAs
Maxisorp 96-well microplates (NUNC) were coated with 5 μg/ml DSPE-PEG (2000) Biotin (Sigma Aldrich). After washing plates with 0.05% CHAPS (Sigma Aldrich) in PBS and blocking the wells with 2% BSA solution, the obtained plasma samples were incubated at 4 different dilutions (1:20, 1:40, 1:80, and 1:160). For the detection of specific PEG-IgG antibodies alkaline phosphatase-conjugated goat-antihuman IgG (Thermo Fisher) was added at 1:2000 dilution. Specific PEG-IgE antibodies were detected by incubating samples first with a 1:3000 dilution of a mouse antihuman IgE followed by adding an alkaline phosphatase-conjugated goat-anti-mouse IgG (Thermo Fischer) antibody at 1:2000 dilution. After a final wash step, substrate buffer containing 1.5 mg/mL Nitrophenylphosphate (NPP, Sigma Aldrich) was added, and plates were read at a wavelength of 405 nm on microplate reader (Berthold Mithras LB940). Specific IgG and IgE Abs to PEG concentrations of each plasma were interpolated from a standard curve created with anti-PEG human-IgG and anti-PEG human-IgE, respectively (Academia Sinica, Taiwan).
Minimum detection cutoffs were determined as OD405 0.2 and OD405 0.4 for PEG IgE and PEG IgG respectively; maximum detection cutoffs were determined as OD405 1.0 and OD405 1.9 for PEG IgE and PEG IgG respectively. High PEG IgG was considered for levels > than OD405 1.5.
2.3 Anti-PEG-IgM ELISA
Maxisorp 96-well microplates (NUNC) were coated with 5 μg/ml DSPE-PEG (2000) Biotin (Sigma Aldrich). After washing plates with 0.05% CHAPS (Sigma Aldrich) in PBS and blocking the wells with 2% BSA solution, the obtained plasma samples were incubated at three different dilutions (1:20, 1:40, and 1:80). Peroxidase-conjugated goat-antihuman IgM (Jackson ImmunoResearch) was added at 1:5000 dilution to detect specific PEG-IgM antibodies. After a final wash step, substrate buffer containing 3,3′,5,5′-Tetramethylbenzidine (TMB) was added, and plates were read at a wavelength of 450 nm on microplate reader (Berthold Mithras LB940). Specific IgM-to-PEG concentrations of each plasma were interpolated from a standard curve created with anti-PEG human-IgM antibody (Academia Sinica, Taiwan).
Minimum detection cutoff was determined as OD450 0.3 for PEG IgM, and maximum detection cutoff was determined as OD450 1.5.
2.4 Complement buffers
Dextrose veronal-buffered saline with divalent cations (DGVB++) (72.7 mM NaCl, 2.47 mM Na-5′-5″ diethyl barbiturate, 1 mM MgCl2, 0.15 mM CaCl2, 2.5% (w/v) dextrose, 0.1% gelatin, pH 7.3–7.4) was prepared following the protocol in the supplement to reference 16.
2.5 Antibody-sensitized sheep cells (EA)
The cells were prepared as previously described in reference 13 with some modifications: Five mL of sheep erythrocytes shipped in Alsever's solution (Colorado Serum Company) were washed 3× with and resuspended in 50 mL of Dulbecco's PBS (DPBS; Sigma, St. Louis, MO) and then divided equally into two centrifuge tubes. 100 μL rabbit anti-sheep RBC/haemolysin (Cedar Lane Labs, catalog # CL9000, resuspended in 1 mL filter-sterilized purified water) was mixed in 50 mL of DPBS. Cells and antibody were separately pre-incubated at 37°C for 10 min. 25 mL of antibody solution was added to each tube of cells with gentle agitation. Cell: antibody mixtures were incubated with gentle rotation at 37°C for 30 min. Cells were washed 2× in 50 mL 10 mM EDTA buffer, 2× in DGVB++, and resuspended in 80 mL of DGVB++ for immediate use. Cell preparations were stored at 4°C.
2.6 Human serum
Pooled human serum was purchased from CompTech (Tyler, TX), divided into 0.1 mL aliquots and stored at −80°C until use.
2.7 Vaccines
BNT162b2 COVID-19 vaccine was obtained from the Barnes-Jewish Hospital/Washington University vaccine clinics and used the same day or stored at 4°C until use. Doxil Control nanoparticle, a PEGylated liposomal vehicle, was purchased from Cat. #300113 (Avanti Polar Lipids, AL) and stored at −80°C until use.
2.8 Vaccine/NP/serum incubation and hemolytic titration
RHA ratios for individual sera are averages of determinations obtained on 3–5 separate days. A value of 1 indicates no detectable vaccine-mediated complement activation while a value near 0 indicates extensive vaccine-mediated complement activation.
2.9 Depletion of IgG from human sera
Sepharose-conjugated Protein A/G (cat# ab193262, Abcam, Cambridge, MA) was used to deplete IgG antibodies from human sera. Protein A/G-Sepharose (150 μL) slurries were washed extensively with PBS 3×, supernatant removed, human serum was added (150 μL) to pelleted beads. Sera were incubated for 1 h at room temperature with end-over-end mixing to allow IgG from serum to bind to the resins. Control sera were processed the same way but without the Sepharose/agarose beads. Following incubation, the beads were pelleted by centrifugation; serum was carefully removed to avoid contamination with beads and used the same day or frozen at −80°C until use. IgG depletion was confirmed by Western blotting.
2.10 PBMC isolation and basophil enrichment
Nonatopic healthy volunteers were recruited to obtain donor basophils, and PBMCs were isolated from fresh heparinized venous blood through centrifugation over Ficoll gradients. Heparinized blood was diluted 1:1 with RPMI-1640 media (Invitrogen, Paisley, United Kingdom), layered on 30% Ficoll-Paque™ PLUS density gradient (GE Healthcare, Uppsala, Sweden), and centrifuged for 25 min at 1136 g at room temperature. PBMC layer was collected, washed, and resuspended in RPMI-1640. Cell viability was determined using trypan blue exclusion.
In experiments where enriched basophils were used, leukocyte-rich plasma was prepared from fresh peripheral blood sample by sedimentation of red blood cells (RBC) through HetaSep™ (StemCell Technologies, Cambridge, UK) using centrifugation. In brief, 1-part HetaSep™ solution was added to 5-parts of fresh whole blood and centrifuged at 90 g for 4 min at room temperature with breaks off. Leukocyte-rich supernatant was harvested and washed to remove platelets. Enrichment of basophil was performed using EasySep™ Human Basophil Isolation kit following the manufacturer's instructions (StemCell Technologies, Cambridge, UK). All purified cells were counted with trypan blue exclusion and checked for purity before processing utilized for downstream processes.
2.11 Stripping and resensitization of basophils
Peripheral blood mononuclear cells (PBMCs) or enriched basophils were isolated from whole blood collected from nonatopic healthy subjects. To strip basophils from their native IgE, cells were treated with 0.01 M (4%) lactic acid-containing buffer (pH 3.9) for 2 min at 4°C. Cells were washed with Ca2+ and Mg2+-free buffer and resensitized with IgE-containing serum for 20 min at 37°C, followed by incubation at 4°C for 30 min.
2.12 Basophil activation test
Basophils resensitized with various serum samples were assessed for their reactivity towards anti-IgE (5 μg/mL), various vaccine components and varying doses of either BNT162b2 or mRNA-1273 vaccine (0.7, 7, and 15 μg/mL) in a 37°C water bath for 30 min. In conditions where C5aRA (75 nM) or C3aRA (1 μM) were tested, cells were pretreated with these antagonist for 30 min at 37°C and washed twice with PBS before stimulated with vaccine. Cells were immunostained with antihuman CD3 (BD Biosciences), CD303, CD294 (CRTh2) (both, Miltenyi Biotech), CD63, C5aR, and C3aR (all from Biolegend unless indicated otherwise). Samples were washed with 2 mL PBS (without Ca2+ and Mg2+) and centrifuged (for 5 min, 400 g) before incubation with Fixable viability stain 780 (FVS780) (Thermo Fisher) for 10 min in the dark. Samples were washed with 2 mL PBS, and cell pellet was resuspended in 150 μL of ice-cold fixative solution (CellFix, BD Biosciences). All samples were acquired on the BD LSR Fortessa X20, and activated cells are phenotyped as those that are CD63+CRTh2+CD303−. Analyses were performed on FlowJo v10.6.1 and validated using unbiased clustering tools viSNE and FlowSOM (Cytobank).
2.13 Single-cell RNA-seq and ATAC-seq
Cryopreserved PBMCs were recovered and counted using trypan blue exclusion and left to incubate at 37°C for 20 min on its own or with autologous sera at a 1:1 ratio. Cells were prepared for nuclei isolation for single-cell multiome ATAC + GEX Sequencing using the manufacturer's instructions (Rev E). Briefly, cells were washed with wash buffer before being exposed to lysis buffer for 1 min at 4°C. Following a final wash, lysed nuclei suspension was diluted in nuclei buffer and counted using trypan blue to determine cell concentration and viability. Single-cell nuclei were then loaded onto a chromium single-cell chip and prepared using the chromium next GEM single-cell multiome ATAC + gene expression Reagent Kit (10× Genomics) according to the manufacturer's instructions to allow encapsulation with barcoded gel beads at a target capture rate of approximately 10,000 individual nuclei per sample. Single-cell RNA-seq and ATAC-seq libraries were prepared for Illumina sequencing according to the manufacturer's instructions. All samples for a given donor were processed simultaneously with the chromium controller and the resulting libraries were prepared in parallel in a single batch. The RNA and protein libraries were sequenced on a NextSeq 2000. A minimum of 25,000 reads per cell or nuclei were sequenced from the RNA and ATAC libraries.
2.14 Preprocessing and analysis of single-cell RNA and ATAC sequencing data
Raw sequencing reads were processed with the Cell Ranger ARC pipeline (10× genomics, v2.0.2) using the GRCh38 reference genome. The resulting RNA counts and ATAC peaks matrices were further processed in R v4.2.247 with the packages Seurat v4.3.048 and Signac v1.9.049 using default parameters (unless otherwise stated or indicated on Figure 6). A more detailed method can be found on the supplementary material.
2.15 Unbiased clustering analyses
Machine learning-driven unbiased analyses (FlowSOM) were performed on flow cytometry dataset using Cytobank. Analysis using viSNE and FlowSOM was performed on the basophil population and cluster setting was set to surface markers CD63, CD203c, C5aR, and C3aR. FCS files from six serum samples for each condition were concatenated to generate a representative dataset (FCSConcat2). All FlowSOM analysis was performed on a predetermined metacluster setting of 12. Star plots generated from FlowSOM allows the identification of two pieces of information: (1) size of the cluster nodes representing population abundance; (2) proportion of pie chart within each cluster node representing the expression of markers. The distance between the nodes is proportional to the dissimilarity of expression patterns of nodes or clusters.
2.16 Correlation analyses
Simple linear regressions were performed to test for correlations within the dataset (RHA, anti-PEG Ig levels, and basophil activity) for each group of patients using Prism 9. The effect of the varying number of days or weeks between blood draws for each patient does not have a significant impact on the results of the dataset as determined based on multiple linear regression performed in Prism 9. Thus, the varying number of days or weeks was not controlled for within the simple linear regression performed on the dataset. To examine the strength of the relationship, Pearson correlation coefficient r was used to calculate p value. Correlations are considered statistically significant if the p value is <.05.
3 RESULTS
3.1 Subhead 1: Characteristics of study participants
A total of 32 adults who received either the BNT162b2 or mRNA-1273 vaccines were enrolled for these studies. Of the individuals who reported allergic reactions to the COVID-19 mRNA vaccines, 19 met the definition of having experienced an immediate allergic reaction as per our prior publication, classified as the allergic group.18 Median interval between vaccination and allergic reaction was 5 min (range 1–120 min); 12/19 participants (63%) had onset within 5 min; 15/19 participants (80%) had onset within 20 min. We also enrolled 13 age-matched individuals who had received COVID-19 mRNA vaccines without allergic reactions, classified as the nonallergic control group. The demographics of all participants are shown in Tables 1 and 2. Four subjects in the allergic group reacted within 120 min of receiving the second dose of the vaccine (but did not react after the first dose), whereas the remaining 15 subjects reacted within 20 min of receiving the first dose of the vaccine.
| Variable | nonallergic N = 13 N (%) | Allergic Reaction N = 19 N (%) |
|---|---|---|
| Age at Enrollment with Median (Range) | 36 (27–79) | 42 (28–70) |
| Sex | ||
| Female | 8 (62) | 17 (89) |
| Male | 5 (38) | 2 (11) |
| Race | ||
| White | 4 (31) | 10 (53) |
| Asian | 7 (54) | 3 (16) |
| Black | 1 (8) | 2 (11) |
| Native American | 1 (8) | 0 (0) |
| Other/Mixed | 0 (0) | 4 (21) |
| Ethnicity | ||
| Not of Hispanic, Latinx, or Spanish origin | 13 (100) | 18 (95) |
| Hispanic, Latinx, Spanish origin | 0 (0) | 1 (5) |
| History of allergies | ||
| Drug allergic history | 2 (15) | 10 (53) |
| Food allergic history | 0 (0) | 3 (16) |
| Latex allergic history | 1 (8) | 0 (0) |
| Environmental allergic history | 2 (15) | 2 (11) |
| Unkown etiology allergic history | 0 (0) | 1 (5) |
| No allergic history | 10 (77) | 8 (42) |
| Days between vaccine and first blood draw, days (median [range]) | 0 (0–45) | 21 (0–86) |
| IgG to PEG, ng/mL-median (range) in first sample | 0 (0–405.21) | 314.8 (0–6903.24) |
| IgM to PEG-median (range) in first sample | 356.35 (0–2716.63) | 203.22 (0–2107.27) |
| IgE to PEG, ng/mL-median (range) in first sample | 0 (0–0) | 0 (0–0) |
| PID | Age | Sex | Race/Ethnicity | Alpha Tryptasemia Testing | Type of vaccine | Hx of allergies | Reaction after dose 1 or 2 | Onset after receipt (min) | Signs and symptoms during the initial reaction | Days Between first dose and blood draw one | IgE levels (ng/mL) at draw one | IgG levels (ng/mL) at draw one | IgM levels (ng/mL) at draw one | Days between first dose and blood draw two | IgE levels (ng/mL) at draw two | IgG levels (ng/mL) at draw two | IgM levels (ng/mL) at draw Two |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A-1 | 20–29 | F | White / Non-Hispanic/Latino | Negative | BNT162b2 | N | Dose 2 | Within 120 min | Redness At Site, Urticaria, Pruritis on leg | 0 Days | 0 | 888.44 | 205.97 | 44 Days | 0 | 987.29 | 190.23 |
| A-2 | 20–29 | F | Black / Non-Hispanic/Latino | Negative | BNT162b2 | N | Dose 2 | Within 120 min | Difficulty Breathing | 0 Days | 0 | 156.62 | 207.64 | 45 days | 0 | 514.86 | 5794.06 |
| A-3 | 30–39 | M | Asian / Non-Hispanic/Latino | Negative | BNT162b2 | N | Dose 2 | Within 120 min | Swelling At Site, Urticaria, Lip Edema | 0 Days | 0 | 626.01 | 203.22 | 42 days | 0 | 901.61 | 261.45 |
| A-4 | 70–79 | M | Asian / Non-Hispanic/Latino | Negative | BNT162b2 | N | Dose 2 | Within 120 min | Redness at Site, Swelling At Site, Urticaria on Tongue and Philthrum | 0 Days | 0 | 491.41 | 0 | 42 days | 0 | 317.11 | 0 |
| A-5 | 50–59 | F | White / Non-Hispanic/Latino | Negative | mRNA-1273 | Drug and Food | Dose 1 | 1 min | Itchiness on Lip, Tachycardia, Dizziness | 20 Days | 0 | 586.02 | 380.71 | 57 days | 0 | 679.07 | 313.25 |
| A-6 | 50–59 | F | White / Non-Hispanic/Latino | Negative | BNT162b2 | Drug | Dose 1 | 2 min | Hypertension, Dizziness | 68 Days | 0 | 1518.63 | 2107.27 | Only one draw | NA | NA | NA |
| A-7 | 30–39 | F | Mixed / Non-Hispanic/Latino | Negative | mRNA-1273 | Drug | Dose 1 | 1 min | Dizziness, Throat Swelling | 34 Days | 0 | 6903.24 | 1349.01 | Only one draw | NA | NA | NA |
| A-8 | 30–39 | F | Mixed / Hispanic/Latino | Negative | BNT162b2 | Drug and Food | Dose 1 | 1 min | Rash, Urticaria on Opposite Arm | 38 Days | 0 | 679.9 | 0 | 74 Days | 0 | 390.32 | 0 |
| A-9 | 30–39 | F | Asian / Non-Hispanic/Latino | Negative | BNT162b2 | Drug | Dose 1 | 1 min | Urticaria on Trunk | 47 Days | 0 | 0 | 359.09 | 78 Days | 0 | 667.56 | 364.23 |
| A-10 | 40–49 | F | White / Non-Hispanic/Latino | Negative | mRNA-1273 | N | Dose 1 | 14 min | Urticaria, Edema Of Tounge | 48 Days | 0 | 198.88 | 181.86 | Only one draw | NA | NA | NA |
| A-11 | 50–59 | F | White / Non-Hispanic/Latino | Not Tested | BNT162b2 | N | Dose 1 | 1 min | Edema, Erythema, Throat Swelling, Tachycardia | 19 Days | 0 | 0 | 109.27 | Only one draw | NA | NA | NA |
| A-12 | 50–59 | F | White / Non-Hispanic/Latino | Not tested | BNT162b2 | Drug | Dose 1 | 1 min | Shortness of Breath, Tachycardia | 17 Days | 0 | 0 | 1658.88 | Only one draw | NA | NA | NA |
| A-13 | 40–49 | F | Mixed / Non-Hispanic/Latino | Not tested | BNT162b2 | N | Dose 1 | 5 min | Dizziness, Hypertension, Chllls, Headache | 21 Days | 0 | 314.8 | 1223.42 | Only one draw | NA | NA | NA |
| A-14 | 40–49 | F | White / Non-Hispanic/Latino | Not tested | mRNA-1273 | Drug | Dose 1 | 20 min | Urticaria, Edema, Lip Swelling, Hives, Shortness of Breath | 12 Days | 0 | 0 | 0 | Only one draw | NA | NA | NA |
| A-15 | 30–39 | F | White / Non-Hispanic/Latino | Not tested | BNT162b2 | Drug | Dose 1 | 5 min | Urticaria, Edema, Throat Swelling, Stridor, Shortness of Breath | 86 Days | 0 | 0 | 466.16 | Only one draw | NA | NA | NA |
| A-16 | 30–39 | F | White / Non-Hispanic/Latino | Not tested | BNT162b2 | Drug and environmental | Dose 1 | 20 min | Facial Edema, Erythema, Hypotension | 33 Days | 0 | 1181.77 | 113.6 | Only one draw | NA | NA | NA |
| A-17 | 50–59 | F | White / Non-Hispanic/Latino | Not tested | BNT162b2 | N | Dose 1 | 3 min | Edema, Tachycardia, Lip edema | 38 Days | 0 | 0 | 131.91 | Only one draw | NA | NA | NA |
| A-18 | 20–29 | F | Mixed / Non-Hispanic/Latino | Not tested | BNT162b2 | Unknown etiology | Dose 1 | 5 min | Urticaria, Pruritis on Chest and Back | 34 Days | 0 | 986.89 | 0 | Only one draw | NA | NA | NA |
| A-19 | 50–59 | F | Black / Non-Hispanic/Latino | Not tested | BNT162b2 | Drug, food, and environmental | Dose 1 | 5 min | Edema, Tongue Swelling, Dizziness | 30 Day | 0 | 0 | 140.19 | Only one draw | NA | NA | NA |
- Abbreviation: NA, Not available.
3.2 Subhead 2: Serum and vaccine-mediated basophil responsiveness
Basophils are key effector cells that play a crucial role in both IgE-dependent and independent allergic reactions. To investigate the underlying mechanism of possible IgE-mediated allergic reactions following BNT162b2 or mRNA-1273 vaccination, we measured the effect of the whole vaccines, along with their ‘inactive’ components (excipients PEG2000 and P80) on basophils using an in-house validated indirect basophil activation assay (iBAT).19, 20 PBMCs or enriched basophils from nonatopic donors were stripped of their native cell-bound surface IgE using 4% lactic acid followed by resensitization with a control serum collected from an individual with an allergy to the grass pollen (GP) antigen Phleum pratense (Phl p) or serum collected from nonallergic and allergic groups.
We tested our iBAT system by testing the effect of stripping and resensitization of basophils with the control GP serum using enriched basophils or PBMCs. Upon stripping of cell-bound surface IgE, we observed a reduction in anti-IgE-mediated basophil activation. This effect was reversed following resensitization with the GP serum in both cell systems (Figure 1A). Furthermore, resensitization of IgE-stripped cells with GP serum increased in the proportion of CD63+ basophils in response to grass pollen allergen Phl p purified protein, confirming the binding of serum grass pollen-specific IgE onto the surface of donor basophils. In the same comparative model of enriched basophils, or PBMCs, we investigated the effect of BNT162b2 vaccine, PEG2000, and P80 on basophil activation following resensitization of cells with serum collected from individuals who have allergies to the vaccine. Vaccine at 15 μg/mL resulted in basophil activation (increase in percentage of CD63+ cells), which was comparable when using either enriched basophils or PBMCs (Figure 1B). In this validation experiment, we performed this side-by-side basophil versus PBMC comparison in five allergic serum samples using two donor basophils. This concentration is similar to that used in previously published BAT assays.18 No basophil activation was observed following stimulation with PEG2000 or P80 alone, indicating serum or cell components were needed for basophil reactivity to occur.
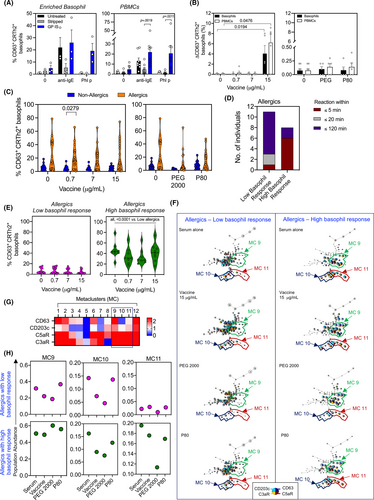
To further investigate the effects of different sera from nonallergic and allergic individuals on basophil activation, we first examined varying concentrations of the COVID-19 mRNA vaccines (BNT162b2 or mRNA-1273) on basophils upon resensitization with PBMC incubation. Overall, we found basophil activation to be generally higher when using sera from allergic compared to the nonallergic group. However, a significant difference in the percentage of CD63+ basophils was only observed at 0.7 μg/mL of the vaccine (p < .05, Figure 1C) while a significant increase in the percentage of CD203cbright basophils was observed across all vaccine concentrations (all, p < .05, Figure S1A). To identify whether the time of onset was associated with basophil activation, we stratified the allergic group into those who developed reaction within 5, 20, or 120 min (Figure 1D). We found that most individuals whose serum induced the highest level of basophil activation (‘high basophil responder’) had a shorter time to allergic reaction development (mostly within 5 min of receiving the vaccine). A significant difference in the percentage of CD63+ and CD203cbright basophils was found in serum-induced basophil activation in the ‘high basophil responders’ compared to ‘low basophil responders’ (Figure 1E and Figure S1B; all, p < .001). There were no differences in BAT activation assays detected between the mRNA vaccines (BNT162b2 or mRNA-1273).
To substantiate and confirm our flow cytometry observations, we used an unbiased machine learning algorithm, FlowSOM, to better characterize cell subsets that may be targeted by the serum, the vaccine, or the vaccine components. FlowSOM analysis allows the unbiased subclassification of cells into metaclusters (MC) of similar phenotypes. In summary, our analysis revealed three specific metaclusters (MC9, MC10, and MC11) that were present in different proportions in ‘low’ and ‘high’ basophil responder individuals (Figure 1F). Heatmap analysis used to illustrate the expression of CD63 and CD203c in each MC demonstrated that MC9 and 11 corresponded to activated basophils that expressed both CD63 and CD203c (Figure 1G). MC10, on the other hand, comprised of activated basophils expressing the CD63lo and CD203cbright phenotypes. Quantification of each MC showed higher abundance in the MC11 following resensitization with serum alone in the ‘high basophil responders’ compared to the ‘low basophil responders’ (Figure 1H). These results suggest that components in certain sera of the ‘high basophil responders’ cohort can elicit MC11, independent of COVID-19 vaccine.
3.3 Subhead 3: Anti-PEG IgE, IgG, and IgM
Specific anti-PEG IgE, IgG, and IgM antibody concentrations in nonallergic and allergic groups were measured by direct ELISA using 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000] (DSPE-PEG(2000)-Biotin) as the antigen source. Absolute concentrations of specific PEG IgE, IgG, and IgM were interpolated from a standard curve. Anti-PEG IgE was undetectable in all subjects (Figure 2A). Thirty-seven percent of the nonallergic group were positive for specific anti-PEG IgG (range 157–405 ng/mL) while 63.2% of allergic individuals harbored specific anti-PEG IgG (range 156–6903 ng/mL) (Figure 2B). The range of anti-PEG IgG antibodies tended to decrease over time (Figure 2B) in allergic individuals, even for those who received a second dose of vaccine. Of note, we also evaluated anti-spike protein IgG antibody responses and found no differences between allergic and nonallergic individuals.21
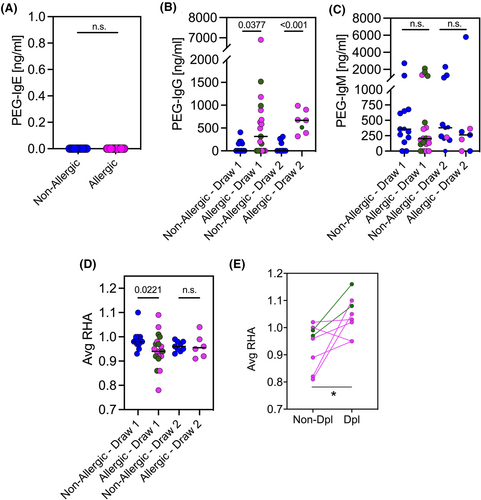
The two groups were stratified according to the timing (i.e., days) of blood collection after the first administration of the vaccine. A significant differences between the nonallergic and allergic groups were noted for specific anti-PEG IgG at both early (Draw 1, p = .0377) and late (Draw 2, p < .0001) blood collection time points (Figure 2B). The percentage of participants who had specific anti-PEG IgM antibodies was similar in nonallergic and allergic groups, 85.7% and 78.9%, respectively. In addition, no significant differences in the median of specific PEG IgM titers could be observed between the groups (Figure 2C).
3.4 Subhead 4: COVID-19 mRNA vaccine-mediated hemolytic activity
To investigate whether CARPA contributes to the COVID-19 mRNA vaccine reactions, we examined the capacity for the vaccines to promote serum complement activation. We used a validated, standard hemolysis assay that measures the serum dilutions that promote complement-dependent lysis of antibody (Ab)-sensitized sheep red blood cells.22 We previously established that pre-incubation of a serum sample with complement-reactive nanoparticles depleted critical complement components and diminished the subsequent hemolytic activity of that serum, indicating that a complement-mediated reaction occurred.22 Herein, the BNT162b2 mRNA vaccine caused little loss of hemolytic activity in this assay when pre-incubated with pooled normal human serum (NHS) (Figure S2A). In contrast, as a positive control, NHS pre-incubated with zymosan or with peroxidase/anti-peroxidase complex (PAP), two strong complement activators, led to significantly diminished residual hemolytic activity (Figure S2A).
Both COVID-19 mRNA vaccines are lipid nanoparticles formulated with different components including PEG-lipids, specifically PEG2000 that could trigger complement activation in those individuals harboring anti-PEG IgG and/or IgM. To test this possibility, we studied the ability of a well characterised rabbit anti-PEG IgG to mediate complement activation in NHS exposed to the BNT162b2 vaccine. We quantified vaccine-mediated complement depletion using the residual hemolytic activity (RHA) metric, which compared the areas under the titration curve obtained with NHS pre-incubated with the vaccine to that of NHS pre-incubated in buffer alone. RHA measures vaccine-mediated complement activation in vitro and is independent of any pre-existing complement activation products. RHA ratios near 1 indicate little or no vaccine-dependent complement activation while RHAs near 0 indicate complete vaccine-mediated depletion of serum complement activity. There was little loss of hemolytic activity occurred when NHS was pre-incubated with either vaccine or anti-PEG IgG (Figure S2B). In contrast, hemolytic activity was depleted when serum was pre-incubated with vaccine and anti-PEG IgG together. As seen in Figure S2B, we found a high degree of correlation between RHAs and exogenously added anti-PEG IgG concentrations (R2 = .9414, p = .0013). Similar correlation was obtained with a Doxil® control nanoparticle, a liposomal formulation that also contains PEG2000 (R2 = .9089, p = .0120, Figure S2C), strongly suggesting that PEG-anti-PEG antibody complex drives this in vitro complement activation in a concentration-dependent manner.
Next, we assayed sera obtained from the two populations, nonallergic and allergic. Sera were incubated with the BNT162b2 vaccine and subjected to the hemolysis assay. We noted significantly higher complement-activation activity (i.e., lower average RHAs) in Draw 1 sera of the allergic population compared with Draw 1 sera of the nonallergic population (p = .0221) (Figure 2D). On the other hand, we noted no significant differences in Draw 2 sera (i.e., similar average RHAs, Figure 2D) between the groups. While the average RHAs of nonallergic sera remained relatively stable between day 0 and day 50 following the first vaccination (Figure S2D), the average RHAs of allergic sera tended to be lower at day 50 (Figure S2E). Although there were striking differences in the anti-PEG IgG levels of nonallergic and allergic individuals (Figure 2B), we found no such correlation between RHAs and anti-PEG IgG (Figure S2F) or anti-PEG IgM levels when considered as a group (Figure S2G). Nonetheless, depletion of total IgG (confirmed by the absence of IgG heavy and light chains on Western blot) from a subgroup of allergic individuals led to higher RHAs (Figure 2E), suggesting that IgG antibody partially contributed to the observed vaccine-mediated complement-depleting activity.
3.5 Subhead 5: Inhibition of C5a and C3a receptors suppressed serum+vaccine-induced basophil responsiveness
Our analyses thus far showed that sera obtained from individuals who had received the BNT162b2 or mRNA-1273 vaccine had varying capacity to activate complement when exposed to BNT162b2 vaccine ex vivo (Figure 2D). Complement activation leads to the generation of anaphylatoxins (i.e., C3a and C5a), which bind to their respective receptors and potently activate innate immune cells such as basophils. First, we quantified the proportion of C3a and C5a receptors (C3aR and C5aR) on the surface of enriched basophils and PBMCs from nonatopic healthy donors and confirmed that C5aR and C3aR were highly expressed, approximately 90% and 80%, respectively, in both of enriched basophils and PBMCs (Figure 1F and Figure 3A). Thus, all analyses involving complement antagonists, hereafter, were performed on PBMCs.
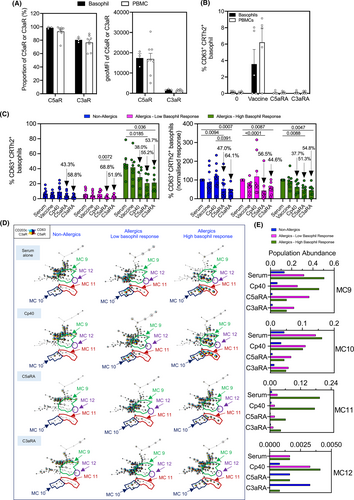
We next investigated the role of complement in basophil activation using two different antagonists: C3aR antagonist (C3aRA) and C5aR antagonist (C5aRA).23, 24 First, we used serum from six allergic individuals to evaluate the effects of C3aRA and C5aRA on vaccine-induced basophil activation. We found that C3aRA and C5aRA, at the concentrations used, both profoundly inhibited vaccine-mediated basophil activation in these test sera (Figure 3B). Next, we examined the effects of C3aRA and C5aRA on basophil activation induced by sera from nonallergic and allergic individuals. When considered as a group, we observed equivalent suppression in serum + vaccine-induced basophil activation following C3aRA and C5aRA pretreatment of sera from both nonallergic and allergic individuals (Figure 3C and Figure S1C,D). When the sera were examined individually, we found a range of suppression by either C3aRA or C5aRA (Figure S1C,D). However, pretreatment with C3aRA and C5aRA combined resulted in no further suppression of vaccine-induced basophil activation (Figure S1E). The fact that we observed suppression of activation in this test system with C3aRA and C5aRA provides strong evidence for complement involvement in serum+vaccine-induced basophil activation. Unbiased clustering analysis using FlowSOM highlighted the same 3 MCs (MC9–11) targeted by C5aRA and C3aRA (Figure 3D). Quantification of population abundance showed that the MCs predominantly inhibited by complement receptor antagonists in nonallergics and low basophil responders were MC9 and MC9/MC10, respectively. Meanwhile, complement receptor antagonists suppressed MC9, MC10, and MC11 in the allergic, high basophil responders (Figure 3E). Interestingly, one particular MC that appeared to be suppressed across all study subjects was MC9, which displayed high expression of CD63 and CD203c, denoting a highly activated state (Figure S3). The high expression of basophil activation markers in MC9 was accompanied by low expression of C5aR and C3aR, which could be due to receptor internalization, further highlighting their potential role in serum-mediated basophil activation.
3.6 Subhead 6: Correlations between RHA:PEG Ig levels and RHA: basophil response
Although we observed low correlation between RHA and anti-PEG Ig levels when all the allergic participants were considered as a group (Figure S2F), we found moderate correlation between RHA and anti-PEG IgG levels when the ‘high basophil responders’ group was examined separately (R2 = .5813, p = .028, Figure 4A). These results suggest that anti-PEG IgG was at least partially responsible for in vitro vaccine-induced complement activation in some of the high basophil responders. On the other hand, there was no correlation between RHA and anti-PEG IgM levels in the ‘high basophil responders’ (R2 = .056, p = .573, Figure 4B). Moreover, we found no or weak correlations between RHA and anti-PEG IgG/IgM in the low basophil responders (Figure S4A–B) and the ‘nonallergics’ (Figure S4E,F). And while RHA, which measures the serum's potential to activate complement ex vivo in the presence of BNT162b2 vaccine, correlates with anti-PEG IgG levels in high basophil responders (Figure 4A), we found no correlation between serum C3a and anti-PEG IgG levels in allergic participants (Figure S4G,H). Moreover, we found no differences in C3a levels between nonallergics and allergics (Figure S4I).
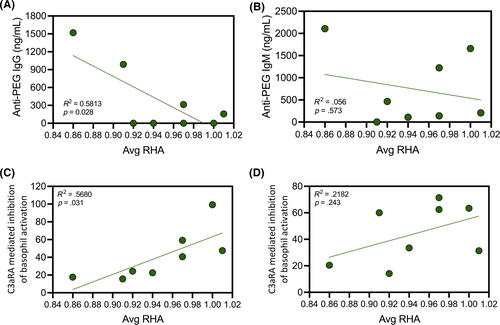
Lastly, we examined the relationship between RHA and basophil response by iBAT. In the ‘high basophil responders’ group, we found that RHA correlated with the degree of basophil suppression by C3aRA (R2 = .5680, p = .031, Figure 4C); the sera with stronger complement-activating activity (i.e., lower RHAs) presaged a more complete suppression of basophil response by C3aRA (lower residual basophil activity). Conversely, the sera with weak complement-activating activity (i.e., higher RHAs) correlated with less suppression of basophil response by C3aRA. Moreover, there was weak correlation between RHA and basophil response in the presence of C5aRA (R2 = .2182, p = .243, Figure 4D), suggesting that the signaling downstream of C3aR and C5aR is nonoverlapping or that the surface structure of the vaccines may not support efficient assembly of the C5 convertase (and C5a release). Additionally, there was no correlation between RHA and basophil response in the presence of C3aRA or C5aRA in the low basophil responders (Figure S4C,D) and the ‘nonallergics’ (Figure S4J,K).
3.7 Subhead 7: Single-cell transcriptomic and ATAC-seq revealed no differential molecular changes pre- and postserum incubation on PBMCs of different patient groups
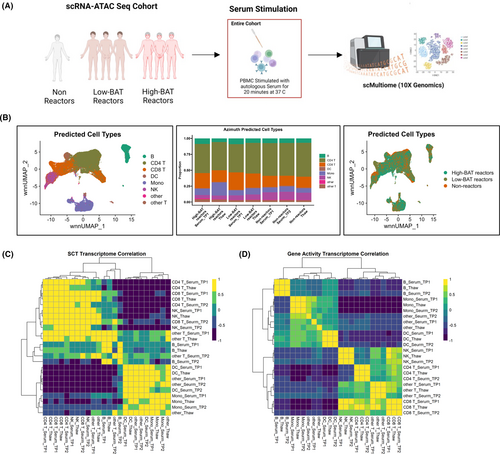
The fact that C3aRA and C5aRA, individually or in combination, were unable to completely suppress vaccine-mediated complement activation suggests that there are other factors contributing to the observed vaccine reactions. Cellular assessments on basophils have highlighted factors within the serum of these study participants that resulted in basophil activation, allowing us to stratify them as ‘high basophil responders’ and ‘low basophil responders’. Here, using single-cell multiome analysis, which simultaneously measure gene expression and chromatin accessibility at a single-cell level, we sought to investigate the differential transcriptomic profile of the high basophil responders, low basophil responders, and nonallergics. A total of seven study participants were investigated at a single-cell level, comprising of three high basophil responders (Hi-BAT reactors), three low basophil responders (low-BAT reactors), and one nonallergic (nonreactor) (Figure 5A). Weighted nearest neigbour (WNN) UMAPs of predicted cell types based on Azimuth classification revealed eight clusters of cells (B cells, CD4+ T cells, CD8+ T cells, dendritic cells, monocytes, natural killer cells, non-CD4 and CD8 T cells, and ‘other’ cells) that were identified within the PBMC population of all study participants included in the single-cell analysis (Figure 5B, left and middle panel). Classifications of these clusters of cells based on the three patient groups demonstrated overlapping pattern and no clear distinction (Figure 5B, right panel). As we were seeing serum-induced basophil activation in the allergic groups, we wanted to understand the molecular mechanisms underlying this. We stimulated PBMCs from study participants with their autologous serum and studied their effect on the transcriptomic and ATAC profile. No transcriptome-wide differences were observed on correlation plots pre- and postserum incubations (Figure 5C,D). Correlation analysis of cell subsets against their serum-treated counterparts did not induce any significant changes both in the RNA expression, as well as chromatin accessibility. Proportion analysis of Azimuth-predicted cell types did not identify any changes in the general cell population pre- and postserum incubation.
3.8 Subhead 8: Single-cell multiome analysis on PBMCs identified contribution of the myeloid compartment on high basophil responders and low basophil responders
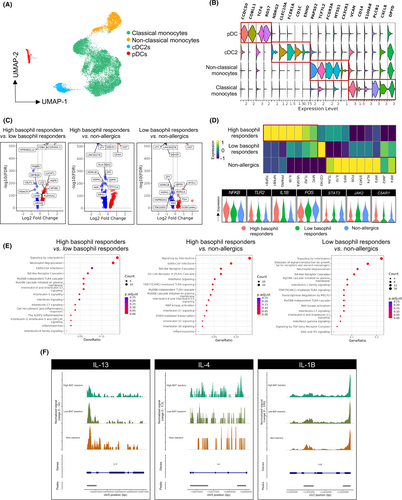
To further understand the molecular mechanisms underlying the response observed in high basophil responders and low basophil responders, we focused our analysis specifically within the innate immune compartment comprising of monocytes and dendritic cells (DCs). A total of 14,846 cells were investigated, of which 13,670 and 1176 were monocytes and DCs, respectively (Figure 6A). Using myeloid cell phenotype annotation, two different clusters of monocytes (CD14+ classical and CD16+ nonclassical monocyte) and two different clusters of DCs (cDC2s and plasmacytoid DCs or pDCs) were identified, with each phenotype of these cell subsets annotated (Figure 6B). Upregulation of common monocyte genes was observed in both the classical (i.e., VCAN, CD14, PLCB1, CXCL8 and DPYD) and nonclassical (i.e., PAPSS2, TCF7L2, FCGR3A, MTSS1 and CX3CR1) subsets. On the other hand, DC subsets express distinct genes such as CCDC50, COBLL1, TCF4 and RGS7 for pDC and NDRG2, CLEC10A, FCER1A, CD1C and ENHO for cDC2 (Figure 6B). Hierarchical analysis was performed to generate a heat map and volcano plot in which a total of 145, 338, and 112 genes were differentially expressed in high basophil responders versus low basophil responders, high basophil responders versus nonallergics, and low basophil responders versus nonallergics, respectively (Figure 6C). A few notable genes were differentially expressed between the three individual groups, indicating differential monocyte activation programs, and include TGFB1, NFKB1, IL1B and TLR2 in high basophil responders, CD163 in low basophil responders, and TLR4, IL-10, IRF3/4/5, STAT3 and JAK2 in nonallergics (Figure 6D). In addition, we observed increased expression of complement receptor C5AR1 in high basophil responders (Figure 6D). Furthermore, pathway enrichment analysis demonstrated vast amount of gene modulation that was associated with signaling by interleukins, SARS-CoV infections, toll-like receptor (TLR) cascade, MyD88 cascade and MAP kinase activation (Figure 6E). To complement the single-cell transcriptomic analysis, chromatin accessibility was assessed for various genes using single-cell ATAC-Seq simultaneously. ATAC-Seq analysis revealed a region on the IL-13 locus that is more accessible in low basophil responders and nonallergics compared to high basophil responders while little differential accessibility of IL-4 (Figure 6F) or IL-10 (Figure S5) was observed in the three different study cohorts. On the other hand, analysis of IL-1B locus shows significantly higher accessibility in high basophil responders and low basophil responders compared with nonallergics (Figure 6F). Taken together, our data indicates that monocytes in high basophil responders are programmed toward M1-like phenotype while nonallergics are polarized toward M2-like phenotype. Low basophil responders, on the other hand, have mixed phenotypes with high CD163 expression (M2),25 high IL-13 chromatin accessibility (M2) combined with high IL-1B chromatin accessibility as well as gene expression (M1). The results suggest that monocytes in vivo are heterogeneous populations of cells that are epigenetically imprinted with different functional programs. In turn, the differential monocyte activation states of the different individuals likely shape the subsequent observed COVID-19 vaccine reactions, especially when considered in the context of basophil activation by vaccine-induced CARPA.
4 DISCUSSION
Allergic reactions to vaccines were initially reported to occur typically at a rate of 1.31 cases per million vaccine doses in large population studies.26 Among those who had immediate allergic reactions to the first vaccination, approximately 0.16% developed severe reactions upon revaccination.27 Allergic reactions are often attributed to inactive ingredients or excipients.5 Both COVID-19 mRNA vaccines contain the excipient PEG2000, which stabilizes the lipid nanoparticle that envelops the mRNA coding for SARS-CoV-2 spike protein. Although the PEG2000 in COVID-19 mRNA vaccines is different from the PEG3350 that is most commonly used in cosmetic and healthcare products, there is suspected pre-existing anti-PEG3350 antibody cross-reactivity to COVID-19 vaccines in reported allergic reactions.5 However, given that only 0.1% individuals in the general population likely harbor anti-PEG IgE antibody28 and none of the individuals who developed allergic reactions to COVID-19 mRNA vaccines in our cohorts exhibited detectable levels of anti-PEG IgE, alternative non-IgE pathways for activating mast cells/basophils were considered in the present study and were investigated at the cellular and molecular level.
Numerous studies have examined the ability of PEGylated lipid nanoparticles to activate the immune system, including complement. The interaction of IgM and IgG with a target surface initiates the complement classical pathway activity, which is then amplified by the complement alternative pathway, a cascade that generates the bioactive proteolytic products C3a and C5a. Our in vitro experiments demonstrated this effect: anti-PEG IgG promoted a robust serum complement response to the BNT162b2 vaccine as well as to a PEGylated Doxil® control lipid nanoparticle. Consistent with a prior literature review, which reported anti-PEG antibody levels in healthy participants ranging from 0.2% to 72%,29 we found that the majority of our participants harbor anti-PEG IgM, IgG or both except for two individuals, N-11 and A-14. Within the allergic group, 17 out of 19 individuals were women, a result that may be partially explained by prior exposure to common over-the-counter products that contain PEG such as lotions and cosmetics.18, 30 Previous studies have also shown that the binding of anti-PEG IgM antibodies to PEGylated liposomes leads to complement activation via the classical pathway and hypersensitivity reactions in animal models.31, 32 However, our results suggest that the presence of anti-PEG IgM alone does not necessarily predict an allergic reaction to the COVID mRNA vaccines, even at high titers. On the other hand, our cohorts exhibited a significant elevations of anti-PEG IgG titers in the group that developed allergic reactions while the nonallergic participants harbored no or low titers of anti-PEG IgG. Moreover, we found a significant correlation (R2 = .5813, p = .028) between serum hemolytic activity (RHA) and anti-PEG IgG levels in the ‘high basophil responders’ but no or weak correlation between RHA and anti-PEG IgM in the low basophil responders and nonallergic groups. There were no observed differences between high basophil responders and low basophil responders concerning RHA and anti-PEG IgG.
We confirmed basophil activation through the presence of CD63+ and CD203cbright surface markers. Although we did observe differences between the percentage of CD63+ basophils and the percentage of CD203cbright basophils across vaccine concentrations, it has previously been shown in the literature that both are markers for basophil activation.33 In addition, the fact that some allergics harbored no anti-PEG antibody and vaccine-induced basophil activation was not completely blocked by C3aRA/C5aRA in some individuals suggests that additional mechanisms likely contribute to the observed allergic reactions to COVID-19 mRNA vaccines. Single-cell RNAseq within the monocyte compartment demonstrated differential gene expression that is associated with downstream TLR signaling pathways, with indication of upregulation of these genes and enrichment of these pathways in the high basophil responders compared to the low basophil responders and nonallergics. We found significant upregulation of M1 activation programs (IL-1B, NF-KB, IL-1A, and TNF) in the high basophil responders and upregulation of M2 activation programs (FOS, JAK2, IL-10, and IL-13) in nonallergics while low basophil responders have mixed M1/M2 activation programs (IL-1B, IL-13, and CD163). These results suggest that the high basophil responders and low basophil responders differ in their functional properties, as reflected in the differential expression of pro- and anti-inflammatory gene expression that is likely modulated by epigenetic mechanisms, as evidenced by differential chromatin accessibility. Additionally, differential expression of CD163, a scavenger receptor that can act as an innate immune sensor to promote inflammation,34, 35 and TLR2 in low basophil responders suggests that these polarized monocytes may release a distinct set of cytokines from high basophil responders upon stimulation, depending on whether the signals are propagated via CD163 or TLR2 or both.
Studies have shown that the ionizable cationic lipids in nanocarriers can directly activate TLR2 and TLR4 on the cell surface,36-38 leading to NF-κB activation and inflammatory mediator release. Whether the COVID-19 mRNA vaccines can directly stimulate CD163 remains to be determined. Complement and TLRs are rapidly activated as a frontline defense and provide a link between innate and adaptive immunity following an infection.39 Complement and TLR crosstalk synergy has been reported at mucosal sites,39 with a study in murine ginginal tissue demonstrating that concomitant activation of C5aR and TLR2 through local injection of their agonists can result in the induction of TNF, IL-1B, IL-6 and IL-17A mRNA and protein.40 In another study, C5a produced during complement activation was shown to contribute towards inflammation through NLRP3 inflammasome, IL-1β, and TNFα release, as well as induction of IL-6 and IL-17.41 These corroborate with our findings in which upregulation of C5aR1 and enrichment of the NLRP3 inflammasome, IL-1, IL-17, and IL-6 signaling were observed in the macrophage compartment of high basophil responders. In addition, the basophil activation and release of cytokines and mediators can further induce the regulation of monocyte polarization and vice versa, contributing to the observed allergic reaction to COVID-19 mRNA vaccines. It has previously been shown by Egawa et al.42 that basophils can drive the differentiation of inflammatory monocytes into M2 macrophages, thereby regulating allergic skin inflammation though not many studies have shown these interaction. In our study, we observed dominance of M1 activation state within the high basophil responders cohort despite a higher level of basophil activation. Whether this discrepancy could be explained by the differential temporal roles displayed by basophils in allergic inflammation, that is, initiatiation of M1 activation state at early stage of allergy followed by attenuation through M2 activation state in the later stages, is yet to be fully elucidated.
In summary, our studies suggest that anti-PEG IgG in the sera of ‘high basophil responders’ may trigger variable degrees of complement activation in the presence of the BNT162b2 vaccine, leading to the release of complement split products that bind to C3aR/C5aR, activating innate immune cells such as basophils to release inflammatory/allergic mediators. In the absence of the vaccine and elevated basophil activation response, our data suggests serum factors from allergic individuals who develop reactions in less than 5 min contribute towards the spontaneous activation. Furthermore, our multiome ATAC-seq analysis revealed differentially polarized monocytes, with M1 phenotype being predominantly observed in the high basophil responders while monocytes from low basophil responders exhibit a mixed M1/M2 phenotype. The monocyte polarization is likely modulated by epigenetic mechanisms43 and determines the subsequent macrophage response upon exposure to COVID-19 vaccine that when combined with CARPA determines the observed allergic phenotype (Figure 7). Of note, BNT162b2 mRNA vaccination has been shown to induce short-term epigenetic changes in innate immune cells.44 However, these changes are short-lived, observed mostly after consecutive vaccination and affecting IFN-stimulated gene expression in monocytes. All subjects in our study received the COVID-19 mRNA vaccines, with all the nonreactors receiving two doses while the majority of the allergics only received one. Moreover, blood samples in the allergics were obtained an average of 21 days after the first vaccination. Thus, we do not believe that monocyte polarization and epigenetic changes in allergics are due solely to COVID-19 mRNA vaccination.
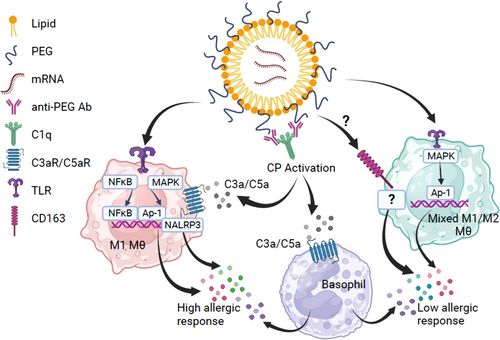
There are likely additional mechanisms. For example, the COVID-19 mRNA vaccines may absorb proteins on its surface once injected in vivo, forming an aggregate or a ‘corona’. This corona comprises proteins such as immunoglobulins, lipoproteins, coagulation factors, and complement proteins,45 which may be directly recognized by basophils/mast cells via the Mas-related G protein-coupled receptor-X246 or different innate immune cells such as neutrophils, leading to histamine and cytokine release. In addition, there still exists the possibility for certain individuals in the population to experience anti-PEG IgE-mediated allergic reactions. Furthermore, it is important to note that healthy donors included in this study possessed detectable IgG but did not experience allergic reactions. This could be explained by differences in the specific IgG subclasses, which we believe should be explored in future studies. Nonetheless, our findings shed new insight into mechanisms of vaccine reactions that may influence ongoing vaccine development, especially as more mRNA vaccines are being developed against novel variants of SARS-CoV-2 and other viruses. The results may also contribute to the development of strategies and guidance to manage subsequent vaccination in individuals with prior allergic responses and help reduce vaccine hesitancy.
4.1 Limitations
There are several limitations to the interpretation of these studies. First, the sera and PBMCs were not collected during the reactions, and thus complement activation and its potential effects on immune cells can only be inferred. Second, the fact that we did not find correlation between IgM levels and complement activating capacity of sera from allergic individuals does not necessarily rule out the contribution of IgM during the reactions since we performed studies using blood collected days to weeks after the reactions. Third, we focused mainly on CARPA and monocyte activation programs as the main mechanisms underlying COVID-19 mRNA vaccine reactions but acknowledge that several alternative mechanisms may be responsible for the observed effects. Fourth, our studies focused on basophils, whereas, in vivo, tissue mast cells may be the main cell type responsible for vaccine reaction and have functions that are partially distinct from basophils. Also, other immune cells such as neutrophils may play a role in these vaccine reactions. The indirect BAT assay uses donor basophils, which are sensitized with IgE to the tested patient. In this study, due to logistical constraint associated with the nature of the COVID-19 pandemic, we were unable to process fresh blood samples on the BAT assay, and thus, iBAT assay offered the best option for us to test their ability to induce basophil activation.
AUTHOR CONTRIBUTIONS
Concept and design: MMS, JAL, DEH, MHS, CTNP, KCN. Acquisition, analysis, or interpretation of data: MMS, JAL, DEH, MHS, CTNP, KCN, AP, MRJ, JMM, ZR, PLK, ASL, JL, DD, IC, MV, AF, PSA, SJG, SDB, BP, MMD, RO, HP, BFG, SRD, AF, IA, AK, MC, LMM, AH, CAA. Obtained funding: KCN, MC, CTNP, DEH. All authors provided intellectual contributions and assisted with writing and editing the manuscript.
ACKNOWLEDGEMENTS
The authors acknowledge and thank the participants who generously shared their time and materials for the purpose of this study. This work would not be possible without the outstanding support of Tarisa Mantia (Washington University) for participant recruitment and Gabija Drazdauskaite (Imperial College London) for the iBAT study. We thank Dr. John D. Lambris (University of Pennsylvania) for the generous gift of C5aRA and Dr. Janos Szebani for helpful scientific insights and critical reading of the manuscript. Figure 7 was created on BioRender.com.
FUNDING INFORMATION
This work was supported by NIH P30AR073752 (C.T.N.P.), NIH UL1TR002345 Pilot award (C.T.N.P., D.E.H.), NIH U54CA260517 (K.C. N, S.C., S.B.S.) and by the Binns Family Foundation (K.C.N.). Infrastructure support and validation of the single-cell multiome methodology for this research was provided by the NIHR Imperial Biomedical Research Centre (BRC) (M.H.S., J.A.L.).
CONFLICT OF INTEREST STATEMENT
All authors declare no competing financial interests.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing was performed as per NIH guidelines. https://sharing.nih.gov/data-management-and-sharing-policy/about-data-management-and-sharing-policies/data-management-and-sharing-policy-overview#after.