A genetic interaction between RAP1 and telomerase reveals an unanticipated role for RAP1 in telomere maintenance
Summary
RAP1 is one of the components of shelterin, the capping complex at chromosome ends or telomeres, although its role in telomere length maintenance and protection has remained elusive. RAP1 also binds subtelomeric repeats and along chromosome arms, where it regulates gene expression and has been shown to function in metabolism control. Telomerase is the enzyme that elongates telomeres, and its deficiency causes a premature aging in humans and mice. We describe an unanticipated genetic interaction between RAP1 and telomerase. While RAP1 deficiency alone does not impact on mouse survival, mice lacking both RAP1 and telomerase show a progressively decreased survival with increasing mouse generations compared to telomerase single mutants. Telomere shortening is more pronounced in Rap1−/− Terc−/− doubly deficient mice than in the single-mutant Terc−/− counterparts, leading to an earlier onset of telomere-induced DNA damage and degenerative pathologies. Telomerase deficiency abolishes obesity and liver steatohepatitis provoked by RAP1 deficiency. Using genomewide ChIP sequencing, we find that progressive telomere shortening owing to telomerase deficiency leads to re-localization of RAP1 from telomeres and subtelomeric regions to extratelomeric sites in a genomewide manner. These findings suggest that although in the presence of sufficient telomere reserve RAP1 is not a key factor for telomere maintenance and protection, it plays a crucial role in the context of telomerase deficiency, thus in agreement with its evolutionary conservation as a telomere component from yeast to humans.
Introduction
Mammalian telomeres are composed of tandem repeats of the TTAGGG sequence bound by a specialized protein complex known as shelterin, which protects chromosome ends and regulates telomerase activity (Takai et al., 2003; de Lange, 2005; Martinez & Blasco, 2010, 2011). Telomere repeats can extend to different lengths in different species, and this length can also vary depending on the developmental stage and cell type within a given species (Flores et al., 2008; Marion et al., 2009). During each cell division cycle, telomeres shorten as a result of the incomplete replication of linear DNA molecules by conventional DNA polymerases, the so-called ‘end-replication problem’ (Watson, 1972; Olovnikov, 1973). Telomerase is a reverse transcriptase (TERT) capable of compensating telomere attrition through de novo addition of TTAGGG repeats onto the chromosome ends using an associated RNA component as template (Terc) (Greider & Blackburn, 1985). Telomerase is expressed in embryonic stem cells and in most adult stem cell compartments; however, this is not sufficient to maintain telomere length, and therefore, telomere shortening takes place with age in most tissues (Harley et al., 1990; Liu et al., 2007; Flores et al., 2008). This progressive telomere shortening has been proposed as one of the molecular hallmarks of aging (Lopez-Otin et al., 2013). Indeed, longevity is progressively decreased upon successive intercrossing of Terc-deficient mice, which is concomitant with a decreased mobilization of adult stem cell populations and premature organ failure. In this regard, Terc-deficient mice show aging-associated pathologies such as alopecia, lower body weight, intestinal atrophy, hair graying, infertility, heart dysfunction, bone marrow aplasia, kidney dysfunction, defective bone marrow, and proliferative defects of neural stem cells (Lee et al., 1998; Herrera et al., 1999; Samper et al., 2002; Blasco, 2005; Garcia-Cao et al., 2006). Telomerase-deficient mice are resistant to cancer, with the only exception of p53-deficient and TRF2-overexpressing genetic backgrounds (Artandi et al., 2000; Blanco et al., 2007).
RAP1 is one of the shelterin complexes that protects the telomeres (for a review, see de Lange, 2005; Martinez & Blasco, 2010, 2011). RAP1 binds to telomeric repeats through its interaction with TRF2 (Li et al., 2000). Human cell lines lacking RAP1 do not show either a DNA damage response (DDR) or telomere fusions except in the absence of TRF2 when RAP1 has a role in inhibition of the NHEJ pathway at telomeres (Sarthy et al., 2009). The role of human RAP1 in telomere length regulation is unclear, while some works have reported no telomere length changes in the absence of RAP1, others have observed telomere lengthening upon both RAP1 knockdown and RAP1 overexpression (Li et al., 2000; O'Connor et al., 2004; Sarthy et al., 2009; Kabir et al., 2014). Mouse cells lacking RAP1 have a moderate increase in fragile telomeres and in telomeric sister chromatid exchange events but are protected against telomere fusions (Martinez et al., 2010; Sfeir et al., 2010). Thus, in contrast to yeast where a crucial role of RAP1 in telomere length regulation and telomere protection has unequivocally been demonstrated (Lustig et al., 1990; Sussel & Shore, 1991; Kyrion et al., 1992; Kanoh & Ishikawa, 2001; Pardo & Marcand, 2005), human and mouse RAP1 does not seem to have an important function in telomere protection. Mammalian RAP1 and their unicellular orthologs have been shown to affect gene expression, and this transcriptional regulatory function has been proposed to reflect the molecular conservation among RAP1 proteins (Kabir et al., 2014).
RAP1 also associates to nontelomeric genomic sites where it has been demonstrated to regulate gene expression (Sarthy et al., 2009; Martinez et al., 2010, 2013; Yang et al., 2011; Yeung et al., 2013). Mice lacking RAP1 are viable but develop obesity, glucose intolerance, and hepatic steatosis. These signs of metabolic disorders are more pronounce in females than in males (Martinez et al., 2013; Yeung et al., 2013). Gene expression profile analyses in liver of adult mice revealed that in the absence of RAP1, several metabolic pathways are remarkably affected (Martinez et al., 2013).
If mammalian RAP1 is dispensable for telomere protection, why has RAP1 been conserved as a mayor telomere-binding protein in mammals? Given its transcriptional regulatory role, it is possible that RAP1 has been conserved as a shelterin component to sense changes in telomeric states and to coordinate the regulation of different signaling pathways involved in different biological functions such as metabolism and DNA repair. To address the potential RAP1 role in the response to short telomeres and to determine whether RAP1 deficiency could rescue the low body weight presented by late-generation telomerase knockout mice, we generated the double knockout Rap1−/−Terc−/− mouse model. We found that mice lacking both RAP1 and telomerase show a progressive decreased survival with increasing mouse generations as compared to telomerase single mutants, demonstrating an unanticipated genetic interaction between RAP1 and telomerase. Telomere shortening was more pronounced in Rap1−/− Terc−/− than in Terc−/− counterparts, leading to an earlier onset of DNA damage and its consequent DDR causing an anticipation of degenerative pathologies in the intestines. In its turn, telomerase deficiency abolishes RAP1-mediated obesity and liver pathologies associated with metabolic syndrome. Cells with shortened telomeres present less amount of telomere-bound RAP1 but show higher numbers of RAP1-bound extratelomeric sites genomewide. Our findings demonstrate that although RAP1 is not a key factor in telomere capping under normal conditions, under certain cellular stresses such as telomerase deficiency, RAP1 exerts an important function for telomere length regulation and protection. These unanticipated findings provide a likely explanation for the fact that RAP1 is the most evolutionarily conserved shelterin.
Results
RAP1 abrogation exacerbates loss of organismal survival associated with short telomeres
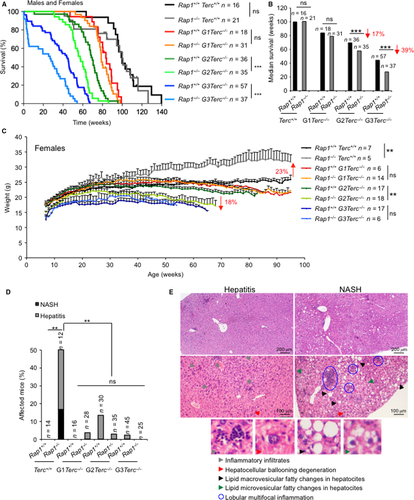
To address a putative role of RAP1 in the organismal response to short telomeres, we generated a Rap1−/−Terc−/− double-mutant mouse by crossing the Rap1−/− with the Terc−/− mice (Blasco et al., 1997; Martinez et al., 2013). The resulting double heterozygous breeding pairs were used to generate first generation (G1) of single Terc−/− and of double Terc−/−Rap1−/−. Large colonies of increasing Terc−/− mouse generations up to the third generation (G3) were generated by intercrossing G1 and G2 Rap1+/−Terc−/− pairs. Survival curves were monitored for the different mouse cohorts under study. As shown in Fig. 1A,B, increasing generations of G1-G3 Terc−/− mice showed a progressive decrease in both median survival and mean lifespan compared to telomerase WT controls, in agreement with previous findings showing a premature loss of mouse survival associated with short telomeres (Herrera et al., 1999; Garcia-Cao et al., 2006). Rap1 deletion did not impact in mouse survival as shown by similar survival curves for single Rap1−/− mutant mice and Rap1+/+ WT controls (Martinez et al., 2013; Yeung et al., 2013). Unexpectedly, simultaneous lack of telomerase and RAP1 in G1-G3 Terc−/−/Rap1−/− mice resulted in decreased survival compared to the RAP1-proficient controls, a difference that reached statistical significance for the late-generations G2 and G3 (Fig. 1A,B). This effect was gender-independent as it was observed both in males and females (Fig. S1A–D, Supporting information). These results suggest that RAP1 deficiency aggravates the telomerase-associated loss of organismal survival.
Telomerase deficiency abolishes RAP1-mediated obesity and liver pathologies
Whole-body Rap1 deletion results in obese mice, being this phenotype more severe in RAP1-deficient females, while telomerase-deficient mice have been shown to present lower body weight than WT mice, most likely owing to intestinal degenerative pathologies (Lee et al., 1998; Herrera et al., 1999; Martinez et al., 2013; Yeung et al., 2013). To assess the effects of double RAP1 and telomerase deficiencies on fat accumulation, we weekly followed body weight from mice of both genders of the different genotypes under study, Rap1+/+Terc+/+, Rap1−/−Terc+/+, G1-G3 Rap1+/+Terc−/−, and G1-G3 Rap1−/−Terc−/− (Figs 1C and S1E, Supporting information). RAP1-deficient females showed a progressive increase in body weight with time from week 8 onwards, reaching an approximately 25% increase in body weight compared to WT females at 1 year of age (Fig. 1C). The increased body weight in Rap1-deficient females was maintained throughout their lifespan (Fig. S1F, Supporting information). Remarkably, female obesity owing to Rap1 deficiency was abrogated in G1-G3 Rap1−/−Terc−/− compound mice and even reversed in G2 Rap1−/−Terc−/− where a significant lower body weight was observed as compared to G2 Rap1+/+Terc−/− females (Fig. 1C). We did not observed significant differences in body weight among the different RAP1-proficient and RAP1-deficient male cohorts (Fig. S1E, Supporting information), in contrast to the moderate increase in body weight previously observed in a different Rap1-deficient mouse cohort (Martinez et al., 2013). This difference in the impact of Rap1 deficiency in male body weight might be explained by slight differences in genetic background associated with generation of the Rap1 Terc compound mouse colony.
We previously reported that Rap1-deficient mice developed liver pathologies reminiscent to those associated with metabolic syndrome, that is, hepatic steatosis and inflammation (Martinez et al., 2013). To address the impact of telomerase deficiency and short telomeres in these pathologies, we performed histopathological analysis of the liver in all the mouse cohorts under study at their time of death. As expected, 50% of Rap1−/−Terc+/+ male and female mice presented liver pathologies that were classified as either severe multifocal hepatitis or as nonalcoholic steatohepatitis (NASH) (Fig. 1D,E) (see Appendix S1, Supporting information). Importantly, no liver pathologies with the exception of some few cases of inflammation associated with aged liver were observed in telomerase-deficient mice regardless of Rap1 genotype (Fig. 1D). These results indicate that RAP1-mediated liver pathologies are abrogated in the context of telomerase-deficient mice.
RAP1 abrogation leads to an earlier onset of aging pathologies associated with short telomeres
To determine the cause of death in the different mouse cohorts, we performed a full histopathological analysis in all the mice at their end point. Atrophies of the intestinal epithelia are the most frequent and life-threatening degenerative lesions associated with short telomeres in G1-G3 Terc−/− mice, which contribute to the early mortality of these mice (Herrera et al., 1999; Garcia-Cao et al., 2006; Martinez et al., 2009a). We classified these intestinal pathologies in mild, medium, or severe according to the pathological findings (see Appendix S1, Supporting information) (Fig. 2A). Severe intestinal atrophy was absent in WT and single Rap1-deficient mice at their time of death (Fig. 2B). In contrast, telomerase-deficient mice showed severe intestinal mucosal lesions characterized by epithelial and glandular atrophy and inflammatory reactions (Fig 2B). Simultaneous telomerase and RAP1 deficiencies further increased the frequency of severe lesions, reaching statistical significance in G2-G3 Rap1−/−Terc−/− compared to the G2-G3 Rap1+/+Terc−/− counterparts (Fig. 2A,B). These observations are in line with the decreased survival observed in G2-G3 Rap1−/−Terc−/− compared with their Rap1+/+ counterparts. Histopathological analysis of kidneys, lungs, spleen, and heart did not reveal any significant differences between G0-G3 Rap1+/+ and G0-G3 Rap1−/− mice (Fig. S2A, Supporting information).
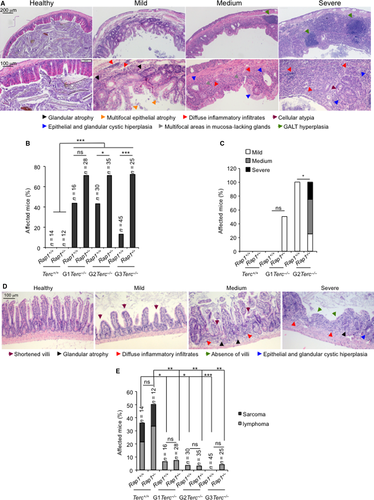
As the above results were obtained at the time of death, to confirm that RAP1 deficiency aggravates degenerative pathologies at the GI track of Terc−/− mice, we sacrificed a group of 4–5 healthy young mice per group before any pathological signs at the age of 40–50 and 22–28 week old in G0 and G1-G3 groups, respectively. Histopathological analysis of the GI track revealed that intestinal pathologies observed in mice showed already generalized degenerative intestinal lesions as well as inflammation, while none of the G2-Rap1+/+Terc−/− mice showed these lesions in a similar age range. Instead, the G2-Rap1+/+Terc−/− animals only showed mild focal lesions. No lesions were observed either in Rap1+/+Terc+/+, Rap1−/−Terc+/+ or in G1-Rap1+/+Terc−/−, while 50% of the G1-Rap1−/−Terc−/− mice already showed mild atrophy (Fig. 2C,D) (see Appendix S1, Supporting information). Thus, RAP1 deficiency accelerates the onset of severe intestinal atrophy in late-generation telomerase-deficient mice. Histopathological analysis of the liver in these mice revealed that 75% of the Rap1−/− mice already present signs of hepatosteatosis, while none of the G2-Rap1−/−Terc−/− showed this pathology (Fig. S2B, Supporting information).
We next analyzed the presence of malignant tumors (i.e. lymphomas and sarcomas) at the time of death of the different mouse cohorts. RAP1 deficiency did not lead to increased incidence of spontaneous malignant tumors (Fig. 2E). Telomerase-deficient mice presented a drastically reduced incidence in malignant tumors at their time of death compared to Terc+/+ mice (Fig. 3E), in agreement with a potent tumor suppressor activity of short telomeres (Gonzalez-Suarez et al., 2000). This tumor protection conferred by telomerase deficiency was not altered in the absence of RAP1 (Fig. 2E).
RAP1 loss leads to a more pronounce telomere shortening in proliferative tissues in TERC-deficient mice
The anticipation and increased severity of degenerative GI pathologies in Terc Rap1 doubly deficient mice suggest a role of RAP1 in telomere maintenance, which is unveiled in the absence of telomerase. In this regard, previous analysis of single RAP1-deficient mice suggested that RAP1 was largely dispensable for telomere maintenance in vivo (Martinez et al., 2010, 2013).
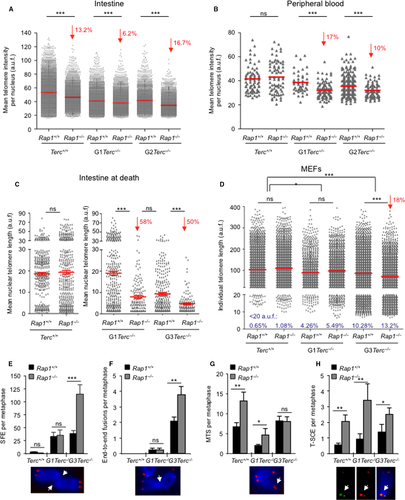
We set to address the impact of Rap1 deletion in telomere length maintenance in vivo in various mouse tissues in the context telomerase deficiency. To this end, we performed telomere quantitative FISH (Q-FISH) analysis on tissue sections of intestine, liver, and of gelatine-embedded peripheral white blood cells (PWBC) from mice of the different genotypes before any pathological signs (same mice as in Fig. 2C,D). Telomere length was moderately but significantly decreased in the RAP1-deficient intestines compared with their RAP1-proficient counterparts, regardless of telomerase status (Fig. 3A). Similarly, we detected a shorter mean telomere length in peripheral white blood cells (PWBC) from G1-G2 Rap1−/−Terc−/− compared with their G1-G2 Rap1+/+Terc−/− counterparts, while no differences in telomere length were detected between Rap1−/−Terc+/+ and Rap1+/+Terc+/+ cohorts (Fig. 3B). In the case of a nonproliferative tissue, such as the liver, however, RAP1 deficiency did not affect mean telomere length either in telomerase WT or in G2 Terc−/− livers (Fig. S2C, Supporting information). These results indicate a role for RAP1 in telomere maintenance in highly proliferative tissues in the context of telomerase deficiency.
We next performed Q-FISH analysis on intestine sections at the time of death in the different mouse cohorts. No significant differences in nuclear telomere length were detected when comparing Rap1+/+Terc+/+ and to Rap1−/−Terc+/+ samples (Fig. 3C). Strikingly, the mean nuclear telomere length of G1-Rap1−/−Terc−/− was dramatically decreased compared with G1-Rap1+/+Terc−/−, reaching values similar to those observed in G3-Rap1+/+Terc−/− mice (Fig. 3C). Similarly, the telomere length of G3-Rap1−/−Terc−/− intestines was significantly reduced compared to G3-Rap1+/+Terc−/−. Thus, mean telomere length in both G1 and G3 Rap1−/−Terc−/− was reduced by 50% compared with G1-G3 Rap1+/+Terc−/− controls. Note that acquisition settings used for microscopy image capture were different in Terc+/+ and Terc−/− samples because the settings used in Terc−/− samples resulted in saturated detention levels in Rap1+/+Terc+/+ and to Rap1−/−Terc+/+ samples (Fig. 3C). Altogether, these data indicate that RAP1 suppresses further telomere shortening associated with tissue homeostasis and aging in the setting of telomerase-deficient mice, thus unveiling an important role for RAP1 in in vivo telomere length regulation.
Finally, we analyzed telomere length in mouse embryonic fibroblasts isolated from the different generation cohorts studied here. To this end, we first established two independent primary embryonic fibroblasts (MEFs) from each genotype under study that we later immortalized by retroviral transduction with SV40 large T (LT) antigen. We carried out Q-FISH on metaphase spreads of immortalized Rap1+/+Terc+/+, Rap1−/−Terc+/+, G1 Rap1+/+Terc−/−, G1 Rap1−/−Terc−/−, G3 Rap1+/+Terc−/−, and G3 Rap1−/−Terc−/− MEFs at passage 10 (Fig. 3D). Deletion of Rap1 did not result in significant differences in mean telomere length between either Rap1+/+Terc+/+ and Rap1−/−Terc+/+ or between G1 Rap1+/+Terc−/− and G1 Rap1−/−Terc−/−, although a modest increase in the percent of critically short telomeres (below percentile 5) was revealed (Fig. 3D). However, G3 Rap1−/−Terc−/− MEFs showed a 18% decrease in telomere fluorescence when compared to their WT controls, which was accompanied by an 3% increase in the frequency of critically short telomeres (below percentile 5). In addition, the number of signal-free ends (SFE) per metaphase was significantly increased by 3-fold in G3 Rap1−/−Terc−/− as compared to their Rap1+/+ counterparts (Fig. 3E). These results further confirm a role for RAP1 in telomere length maintenance in the setting of telomerase deficiency.
RAP1 abrogation induces a more severe telomere uncapping in late-generation Terc-deficient MEFs
Our findings indicate that mice and MEFs lacking RAP1 experience a faster and enhanced telomere shortening in the absence of telomerase. However, a putative role for RAP1 in facilitating telomerase action on chromosome ends is ruled out by our observation that RAP1-mediated telomere erosion is particularly evident in a telomerase-deficient background. Thus, the role of RAP1 in telomere length homeostasis must rather be due to a protective function that is dispensable when telomeres are long but becomes relevant when telomeres are short.
To address this, we performed cytogenetics analysis on metaphase spreads of the immortalized MEFs described above (Fig. 3F–G). In agreement with higher numbers of SFE, we also observed a 2-fold increase in the number of end-to-end fusion events per metaphase in G3 Rap1−/−Terc−/− compared with G3 Rap1+/+Terc−/− MEFs, while no differences were detected between Rap1−/−Terc+/+ and Rap1+/+Terc+/+ or between G1 Rap1−/−Terc−/− and G1 Rap1+/+Terc−/− MEFs (Fig. 3F). As previously reported by us (Martinez et al., 2010), Rap1 deletion induces a moderate increase in the number of multitelomeric signals (MTS), a telomere aberration that is indicative of telomere fragility (Martinez et al., 2009b), both in Terc+/+ and in G1-Terc−/− MEFs. This effect was abolished in G3-Terc−/− probably due to the presence of high numbers of SFE (Fig. 3G). In accordance with a RAP1 role in prevention of homologous recombination at telomeres (Martinez et al., 2010; Sfeir et al., 2010), we also detected a significant increase in sister telomere recombination (T-SCE) frequencies in (G0-G1-G3) Rap1-deleted MEFs compared to Rap1 WT controls as determined by the CO-FISH technique (Fig. 3H).
RAP1 loss triggers DNA damage and accelerates the DDR onset in telomerase-deficient mice
Critically short and uncapped telomeres are recognized as DNA double-strand breaks and, as such, are bound by γH2AX (Hao et al., 2004). In turn, telomere damage elicits a DDR characterized by activation of p53 and the cell cycle inhibitor p21 that leads to cessation of cellular proliferation and cell death, ultimately causing tissue failure and various age-related pathologies (Flores et al., 2005; Martinez & Blasco, 2010).
We addressed whether the earlier onset of GI atrophy in G2-Rap1−/−Terc−/− was associated with an earlier induction of DNA damage and its associated DDR. We quantify the number of γH2AX-, p53-, and p21-positive cells in the intestine of mice of the different genotypes under study. At their time of death, we did not see significant differences in p53 and p21 levels in the GI tract between different Rap1+/+ and Rap1−/− cohorts (Fig. 4A,B), possibly owing to the fact that these mice already had overt intestinal pathologies. Thus, we analyzed these markers at an earlier time in asymptomatic mice of 20–50 weeks of age (Fig. 4C–E). We did not find significant differences in the numbers of γH2AX-, p53-, and p21-positive cells between Rap1+/+Terc+/+ and Rap1−/−Terc+/+. However, late-generation G2-Rap1−/−Terc+/+ mice presented a higher DNA burden and increased p53 and p21 levels compared to G2-Rap1+/+Terc+/+ (Fig. 4C–E). Furthermore, G2-Rap1−/−Terc−/− intestines showed higher number of apoptotic cells (positive for active caspase 3) and reduced proliferation (Ki67-positive cells) compared to G2-Rap1+/+Terc−/− (Fig. 4F,G).
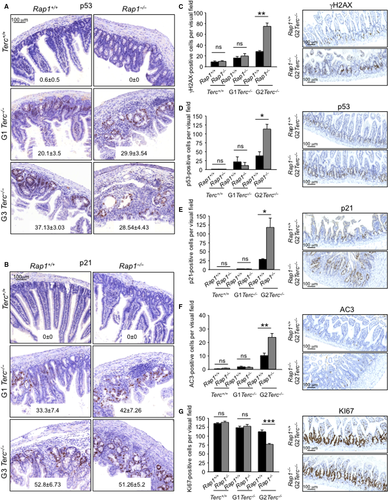
Altogether, these results demonstrate that Rap1 deletion in the context of telomerase deficiency anticipates the induction of p53 and p21 within tissues, concomitant with increased apoptosis and decreased proliferation rates in the GI track of late-generation telomerase-deficient mice, which in turn is accompanied by an anticipated onset of degenerative pathologies and earlier death.
Increased RAP1 genomic occupancy with decreasing telomere length
Recruitment of RAP1 to telomeres occurs through interactions with TRF2, and therefore depended on the number of TRF2 molecules bound to telomeric DNA (Li et al., 2000; Arat & Griffith, 2012; Bandaria et al., 2016). Mammalian RAP1 also binds nontelomeric genomic sites where it regulates gene expression (Sarthy et al., 2009; Martinez et al., 2010, 2013; Yang et al., 2011; Yeung et al., 2013). We hypothesize here that telomere length could affect the ratio between telomere-bound RAP1 and genomic DNA-bound RAP1 that is available to perform its transcriptional regulatory function.
To address this, we first studied whether the total cellular RAP1 level varied with telomere shortening. To do this, we analyzed RAP1 protein levels by Western blot analysis with whole cell extract from immortalized Rap1+/+Terc+/+, Rap1−/−Terc+/+, G1 Rap1+/+Terc−/−, G1 Rap1−/−Terc−/−, G3 Rap1+/+Terc−/−, and G3 Rap1−/−Terc−/− MEFs. We found that progressively shorter telomeres did not affect total RAP1 protein level (Fig. S3A, Supporting information). We then determined the RAP1 protein levels present at telomere foci by performing double immunofluorescence (IF) with RAP1 and TRF1 antibodies. We used Rap1−/−Terc+/+ cells as negative control for RAP1 IF (Fig. 5A–C). We found a significant and progressive decrease in both TRF1 and RAP1 at telomere foci with decreasing telomere length in G1-G3 telomerase-deficient cells (Fig. 5A,B). These results suggest that telomere shortening owing to telomerase deficiency results in decreased RAP1 levels at telomere foci.
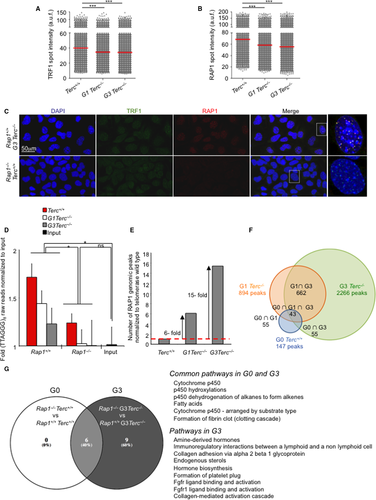
To determine whether the RAP1 genomic chromatin-binding peaks also changed with telomere shortening, we performed a chromatin immunoprecipitation sequencing (ChIP-seq) assay using the same immortalized MEFs (passage 10) used for the IF analysis (Fig 5A–C). The (G0-G1-G3) Rap1-null MEFs were processed in parallel to validate the specificity of RAP1-associated peaks obtained in Rap1 WT MEFs. Over 22 million of raw 50-bp reads were collected for each sample (Table S1, Supporting information). First, we analyzed the representation of the telomeric repeat TTAGGG in the ChIP-seq raw reads of (G0-G1-G3) Rap1+/+ vs. (G0-G1-G3) Rap1−/− samples before genome alignment. We found a strong overrepresentation of 50-bp sequences containing the (TTAGGG)8 repeat in WT Rap1 ChIP-seq as compared to Rap1-null controls, which showed similar levels to input DNA (Fig. 5D). In addition, we observed a progressively reduced number of telomeric reads in successive generations G1-G3 Rap1+/+Terc−/− compared with Rap1+/+Terc+/+ samples, thus confirming that the level of telomere-bound RAP1 is decreased with telomere shortening, in agreement with the RAP1 IF data (Fig. 5B).
Identification of RAP1-bound genomic binding sites revealed 147, 894 and 2,266 nontelomeric peaks in the comparison between the Rap1 WT and the Rap1-null ChIP-seq G0, G1, and G3 datasets, respectively, at 10E-03 P-value cutoff (GEO accession number, GSE79996). Thus, the number of RAP1 genomic binding peaks was progressively increased in cells with progressively shorter telomeres, 6- and 15-fold in G1 and G3 Terc−/− higher than in Terc+/+ WT MEFs, respectively (Fig. 5E). In addition, approximately 40% of the G0 peaks were also found in G1 and G3, and 75% of the peaks identified in G1 were also detected in G3 (Fig. 5F). A total of 1,604 new RAP1 genomic biding sites were observed in G3 (Fig. 5F). Altogether, these results demonstrate that telomere length affects the ratio between telomere-bound RAP1 and genomic DNA-bound RAP1, leading to decreased RAP1 at telomeres and increased RAP1 occupancy at the chromosome arms.
We also confirmed our previous findings (Martinez et al., 2010) that the RAP1 peak density distribution in telomerase-proficient MEFs was progressively decreasing along the chromosome arms toward the centromere, presenting the highest density at the subtelomeric region (Fig. S3B, Supporting information). Interestingly, this trend was inverted in G1 and G3 telomerase-deficient MEFs that present a more evenly distributed peak density along the chromosome arms (Fig. S3B, Supporting information). These findings indicate that telomere shortening not only results in lower RAP1 binding at telomeres but also at subtelomeric sites.
Effects of shortened telomeres on RAP1-mediated transcription
To address whether the increased genomic RAP1 occupancy observed in G1-G3 Terc−/− cells resulted in transcriptional changes, we carried out gene expression analysis by RNA sequencing (RNAseq) analysis from the same immortalized MEFs used for ChIP-seq above. The results have been deposited in GEO (GEO accession number, GSE79996). Then, we performed GSEA analysis in G0-G3 Rap1−/− MEFs (Tables S2 and S3, Supporting information, Fig. 5G). GSEA revealed significant downregulation of pathways involved in lipid metabolism in G0-G3 Rap1−/− cells, being the number of downregulated pathways higher in G3 Rap1−/− than in G0 Rap1−/−. Thus, in G3 Rap1−/− MEFs, pathways related to hormone biosynthesis, adhesion, and fibroblast growth factor receptor were downregulated as compared to RAP1-proficient counterparts. These results are in agreement with the proposed role of RAP1 in the transcriptional activation of genes involved in fatty acid metabolism (Martinez et al., 2010) and with the increased numbers of RAP1-genomic binding sites in G3 Rap1+/+ as compared to Rap1+/+ (Fig. 5E–G). Interestingly, GSEA analysis did not reveal any upregulated pathways in G3 Rap1−/− cells, while several pathways related to cell proliferation, extension of telomere, DNA repair, and carbohydrate–nucleid acid—amino acid metabolism were significantly upregulated in G0 Rap1−/− (Table S3, Supporting information). This observation likely reflects that Rap1 deletion induces a basal state of telomere uncapping that is being repaired by different DNA damage repair pathways, that is, homologous recombination, ATM signaling, and GG-NER. The fact that no differences in the activation of these pathways were detected between G3 Rap1+/+ and G3 Rap1−/− may indicate that in the context of critically short telomeres (G3), both Rap1+/+ and Rap1−/− MEFs present dysfunctional telomeres and therefore similarly activate these DNA damage repair-related pathways. Interestingly, 0.44% of the genes whose expression was altered > 1.5 fold in G0 Rap1−/− presented a RAP1-genomic binding peak while this figure was increased to 2.3% in G3 Rap1−/−, consistent with a higher number of RAP1-binding peaks G3 Rap1+/+ than in G0 Rap1+/+ MEFs (Fig. 5F).
To explore the biological pathways affected by Rap1 deletion, we performed functional analysis with the significantly deregulated genes (FDR < 0.05) between G0-G1-G3 Rap1−/− cells compared to G0-G1-G3 Rap1+/+ cells. For this, we used the Ingenuity software to find disorders and disease groups that were significantly affected (FDR < 0.05) in the three groups (Fig. S4, Supporting information). We found similar biological functions related with disease in the three groups. We did not detect deregulation of any particular disorder/disease in cells with shortened telomeres compared to telomerase-proficient groups that could underline the reduced survival observed in G3 Rap1−/− mice. These results suggest that reduced survival in the Rap1 Terc doubly deficient mice is mainly due to RAP1 function in telomere homeostasis.
Discussion
Here, we demonstrate that RAP1 exerts an important role in telomere maintenance and telomere protection in the context of telomerase deficiency. In line with this, mice lacking both RAP1 and telomerase show a progressive decreased survival with increasing mouse generations compared with telomerase single mutants.
In particular, we observe that telomere erosion due to telomerase deficiency is enhanced in the absence of RAP1, leading to an earlier onset of DNA damage and its consequent DDR (Fig. 6). As a consequence, we observe an accelerated onset and progression of degenerative pathologies (i.e. GI atrophies) in G3 Rap1−/−Terc−/− compared to G3 Rap1+/+Terc−/−, thus leading to shorter mouse survival. These findings unveil a previously unnoticed role for RAP1 in telomere maintenance and tissue homeostasis. Importantly, we observed the most drastic pathological effects of RAP1 deficiency in the context of telomerase deficiency in proliferative tissues such as the intestinal tract, but not in the low-proliferating tissues such as liver, lung, spleen, heart, or kidneys, suggesting that RAP1 is particularly important to maintain homeostasis of the highly proliferative tissues. In fact, RAP1-mediated effects on telomere shortening are only observed in proliferative tissues such as intestines and blood indicating a cell-/tissue-specific RAP1 effect. The mechanisms underlying the accelerated telomere shortening could be explained by a faster cycling rate and/or increased ROS in Rap1−/−Terc−/− as compared to Rap1+/+Terc−/− intestinal cells. These results are in agreement with recent findings supporting that interaction between RAP1 and TRF2 is important for telomere function and chromosome-end protection (Arat and Griffith 2012; Rai et al., 2016). In particular, interaction between RAP1 and TRF2 significantly increases the binding affinity of the complex for double (ds) telomeric repeats and for telomeric 3′ ends (Arat et al. 2012). In addition, the TRF2/RAP1 complex has higher ability to remodel telomeric DNA compared to either component alone. On long telomeres, the TRF2-RAP1 complex can initiate t-loop formation and prevention of the DDR. Similarly, on short telomeres where the t-loop cannot be formed, the TRF2/RAP1 complex can still bind to the ds-ss telomere junction due its high affinity for this structure and suppress the unnecessary DDR by blocking the end and making it less accessible to DNA repair machinery and thereby exerting a protective role (Arat et al. 2012). Thus, our results could be in agreement with a model in which t-loop formation of critically short telomeres after replication is further compromised in RAP1-depleted cells, being more susceptible to telomere degradation by nucleases. Indeed, RAP1 was recently reported to protect from end-to-end fusions in cells with topologically compromised telomeric DNA with significantly reduced cellular t-loop content (Benarroch-Popivker et al., 2016). In agreement with this, G3 Rap1−/− cells present higher incidence of end-to-end fusions (see Fig. 3F).
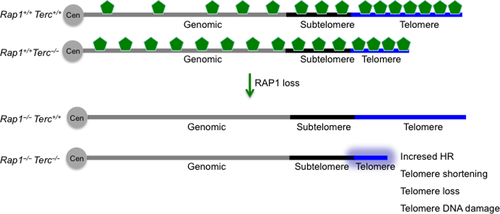
RAP1 has also been shown to be essential to protect telomeres from homology-directed repair as its loss induces telomeric sister chromatin recombination (Martinez et al., 2010; Sfeir et al., 2010; Rai et al., 2016). Here, we confirm a moderate increase in telomeric homologous recombination (HR) in the absence of RAP1 regardless of the underlying telomere length, supporting an antirec function of RAP1. Indeed, it has recently been shown that RAP1 cooperates with the basic domain of TRF2 (TRF2B) to repress the HR factors PARP1 and SLX4 localization to telomeres. In the absence of RAP1 and TRF2B, PARP1 and SLX4 promote telomere resection that results in telomere loss and the generation of telomere-free chromosome fusions mediated by the HR pathway (Rai et al., 2016). Thus, it is tempting to speculate that the enhanced telomere shortening observed in telomerase-deficient cells lacking RAP1 is likely due to increased telomeric HR in the absence of RAP1 (Fig. 6). In addition, it is also possible that telomerase and RAP1 cooperate to protect telomeres from degradation during replication of telomeric DNA tracts through the antirec properties of RAP1.
We also found that RAP1 deficiency did not abolish the tumor suppressor role of short telomeres and telomerase deficiency (Greenberg et al., 1999; Gonzalez-Suarez et al., 2000). This observation is particularly interesting in light of a previous report suggesting a tumor suppressor role for RAP1 via modulation of the NF-κB-mediated pathway (Teo et al., 2010). However, our results clearly show that RAP1 deficiency does not affect spontaneous cancer incidence throughout the lifespan of both telomerase-deficient and telomerase WT mice (Martinez et al., 2013).
RAP1 protein does not contain a DNA-binding domain (Li et al., 2000), and therefore, it is proposed to interact with yet unknown partners to accomplish its transcriptional function (Martinez et al., 2010). However, human RAP1 can bind in vitro to DNA with a preference for double-strand–single-strand junction in a sequence-independent manner (Arat et al. 2012), thus raising the possibility that direct RAP1 binding to DNA could at least partially account for nontelomeric function at internal sites on the chromosomes. The recruitment of RAP1 to the telomeres is accomplished through interactions with the shelterin component TRF2 and is thereby dependent on the number of TRF2 molecules bound to the telomeric DNA repeats, which in turn is proportional to the length of telomeres (Li et al., 2000; Arat & Griffith, 2012; Bandaria et al., 2016). Telomere length is highly dynamic and does vary among different cell types and along aging (Harley et al., 1990; Liu et al., 2007; Flores et al., 2008). Thus, it is therefore conceivable that telomere length would affect the ratio between telomere-bound RAP1 and genomic DNA-bound RAP1 that is available to perform its transcriptional regulatory function. Indeed, in telomerase-deficient senescent yeast, the levels of Rap1 at telomeres decrease concomitantly with an increase in Rap1 occupancy at extratelomeric sites already bound in nonsenescent cells as well as at other newly identified genomic sites denoted as new Rap1 targets at senescence (Platt et al., 2013). In addition, Rap1 delocalization contributes to the transcriptional changes observed in senescent cells and it was suggested that its release from telomere foci is a coordinated genomic response to telomere shortening (Platt et al., 2013). In agreement with this notion, here, we find that short telomeres owing to telomerase deficiency result in decreased RAP1 levels at telomeres as determined by whole-genome ChIP sequencing, at the same time that increases RAP1 peaks at extratelomeric genomic sites in cells (Fig. 6). We also find that the known RAP1 enrichment at subtelomeric sites is lost as telomeres shorten with increasing mouse generations in the absence of telomerase, thus suggesting potential effects in subtelomeric gene silencing (Fig. 6). Accordingly, gene expression profiling in RAP1-proficient and RAP1-deficient G3-Terc−/− MEFs revealed a moderate higher numbers of deregulated pathways in the absence of RAP1. These pathways were related to hormone biosynthesis, adhesion, and fibroblast growth factor receptor. Our data support that RAP1-mediated effects on the survival of telomerase-deficient mice are through its function in telomere homeostasis. However, given that transcriptional profiling in this work was performed in SV40-LT immortalized MEFs, affected in many different pathways including p53 and RB, we cannot rule out that decreased survival in Rap1−/−Terc−/− compound mice could also partially be attributed to RAP1 role in transcriptional regulation. Future work aimed to analyze the RAP1-mediated transcriptional effects in different tissue types of late-generation telomerase-deficient mice is needed to ultimately unveil RAP1 role in the survival of telomerase-deficient mice.
The deleterious in vivo effects of single Rap1 deletion are manifested in the liver and white fat tissues resulting in obesity and hepatic steatosis (Martinez et al., 2013; Yeung et al., 2013). However, telomerase deficiency in combination with RAP1 depletion abrogates the obese phenotype and liver pathologies observed in Rap1 single knockout mice. One likely explanation to this observation is that the severe intestinal degenerative pathologies induced by telomerase deficiency impede proper nutrient absorption at the GI track, thereby inhibiting lipid accumulation. Alternatively, few critically short telomeres in liver cells as a consequence of telomerase deficiency could trigger a metabolic program counteracting the one induced by RAP1 loss.
Our findings demonstrate that although RAP1 is not a key factor in telomere capping under conditions of a sufficient telomere reserve, in the settings of telomerase deficiency, RAP1 exerts an important function in telomere protection and telomere length maintenance, which in turn is important to delay the appearance of degenerative diseases and for organismal longevity. These previously uncovered findings explain its evolutionary conservation as a shelterin component in mammalian cells.
Experimental procedures
Generation of Rap1 Terc double knockout mice
Rap1Terc knockout mice, Rap1−/−Terc−/−, were generated by crossing Rap1−/− (C57BL6/129SV; 90%: 10%) (Martinez et al., 2013) with Terc−/− mice (C57BL6; 100%) (Blasco et al., 1997). The resulting double heterozygous breeding pairs were used to generate G1 of single Terc−/− and of double Rap1−/−Terc−/−. Mouse colonies of successive generations, G2 and G3, were generated by intercrossing G1 and G2 Rap1+/−Terc−/− pairs, respectively. All mice were generated and maintained at Animal Facility of the Spanish National Cancer Research Centre (CNIO) under specific pathogen-free conditions in accordance with the recommendation of the Federation of European Laboratory Animal Science Associations.
Acknowledgment
We are indebted to R. Serrano for animal care and to D. Megias for microscopy analysis.
Author contributions
MAB and PM designed experiments and wrote the manuscript. PM performed the experiments. GG-L and DP performed de RNAseq and ChIPseq analysis. JMF performed the pathology analyses.
Funding
Research in the Blasco laboratory is funded by Spanish Ministry of Economy and Competitiveness (MINECO and FEDER) Project RETOS (SAF2013-45111-R), the European Research Council (ERC) Project TEL STEM CELL (ERC-2008-AdG/232854), and Fundación Botín.
Conflict of interest
None declared.