Identification of differentially expressed circular RNAs associated with thymoma
Qingjun Wu and Xuanmei Luo contributed equally to this work.
Funding information: Beijing Dongcheng District Outstanding Talent Funding, Grant/Award Number: 2019WJGW-10-01; Beijing Hospital Nova Project, Grant/Award Numbers: BJ-2018-137, BJ-2019-149; CAMS Innovation Fund for Medical Sciences, Grant/Award Number: 2018-I2M-1-002; National Natural Science Foundation of China, Grant/Award Numbers: 81870048, 81871107; National Science and Technology Major Project for Significant New Drugs Creation, Grant/Award Number: 2017ZX09304026
Abstract
Background
Thymomas and thymic carcinomas are the most common tumor types among anterior mediastinal lesions. However, the relationship between molecular aberrations and thymoma patients are poorly understood, especially abnormal changes in the expression profiles of circRNAs. The purpose of the present study was to investigate the expression profiles of circRNAs in thymoma patients and their possible roles in the pathogenesis of thymoma.
Methods
Diseased tissues and surrounding normal thymic tissues in two thymoma patients were collected for circRNA sequencing. The top four upregulated circRNAs were selected as candidates and further validated with RT-PCR in 20 thymoma patients. Gene ontology and signal transduction network analyses of circRNA-related mRNAs were performed to analyze the functional properties. Survival analysis of their parental genes were also carried out to evaluate the clinical value of differentially expressed circRNA.
Results
A total of 73 circRNAs were differentially expressed in thymoma tissues using high-throughput sequencing. Among these circRNAs, hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 were significantly upregulated in thymoma tissues compared with normal thymic tissues. We identified that these four circRNA-related mRNAs were involved in cell–cell adhesion, MAPK pathways, and TNF pathway, which may contribute to the pathological immune disorder in thymoma. Finally, we also found that SCAP (hsa_circ_0007291 parental gene) and AFF2 (hsa_circ_0001947 parental gene) were all significantly related with progression-free survival (PFS) of thymoma patients in a Kaplan-Meier plot (p-value <0.05).
Conclusions
The expression levels of hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 were significantly upregulated and positively correlated with immune imbalance in thymoma patients.
INTRODUCTION
Thymoma is a neoplasm with complex histological and biological behaviors that often occurs in the anterior mediastinum.1 Patients with thymomas are often asymptomatic at the initial stage, and they are often unintentionally found during physical examination. Chest imaging examination usually shows a round or lobulated tumor in the anterior mediastinum.2 There are no effective preventive measures for thymoma, but early diagnosis and early treatment are keys to disease control. Thymomas should be surgically removed as soon as possible, and pathological biopsies used to guide postoperative treatment.3
The etiology of thymoma remains to be fully elucidated, but may be related to genetic inheritance. In addition, the thymus is the main place for T lymphocytes to mature and has an indispensable effect on cellular immunity. Therefore, the global gene expression profile of thymic tissue in thymoma patients, especially the immune-related expression profile, needs to be further studied.
CircRNAs (circular RNA molecules) are a kind of non-coding RNA molecule that do not have a 5′- terminal hat and a 3′- terminal poly (A) tail and form a ring structure with covalent bonds. Most circRNA are derived from exons and a few are formed by direct cyclization of introns.4 Recent studies have shown that circRNA molecules are rich in miRNA response elements (MRE) ,and act as an miRNA sponge to indirectly regulates the expression of target genes downstream of miRNA, which is called the competitive endogenous RNA (ceRNA) mechanism.5 Due to the spatiotemporal specificity and disease specificity of expression, circRNAs are expected to have importance in normal development of tissues but also in disease pathogenesis. A number of investigations have demonstrated that circRNAs are associated with several types of cancer, such as gastric cancer, papillary thyroid cancer, hepatocellular carcinoma, and malignant melanoma.6-9
In the present study, RNA sequencing (RNAseq) was performed to detect the expression of circRNAs, and their expressions were further validated in thymoma patients. The functional analysis of circRNAs-related miRNAs and their regulated target genes revealed the immunomodulatory role of circRNAs in the pathogenesis of thymoma.
METHODS
Thymoma patients and thymus tissue samples
Clinical specimens were collected from 22 patients who underwent thymoma resection in Beijing Hospital from January 2018 to December 2019. All samples were diagnosed as type B2 thymoma according to the World Health Organization criteria. The non-neoplastic thymic tissue adjacent to the tumor tissue served as the normal control. None of the patients received radiotherapy, chemotherapy or other treatment before the operation. The study was approved by the ethics committee of Beijing Hospital (2020BJYYEC-007-02), and all the subjects signed informed consent forms.
RNA extraction and RNA-seq
Total RNA was extracted from liquid nitrogen frozen thymoma tissue and adjacent normal tissue by the Trizol method (Invitrogen, Thermo Fisher Scientific). The quality of RNA was detected by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific), and the integrity of RNA was verified by gel electrophoresis. After removing rRNA and linear RNA, random primers were amplified and reverse transcribed into fluorescence-labeled cDNA to complete the preparation of the whole library. The constructed library was sequenced with Illumina HiSeq cDNA 2000. The read quality was filtered through a 4-base sliding window with an average quality threshold of 15, while reads with a length of less than 28 were discarded. The filtered reads were mapped to the human hg38 genome by aligner software STAR.10 This part of the work was entrusted to Annoroad Gene Institute (Beijing, China).
Quantitative real-time PCR
A total of 5 μg total RNA was digested with Rnase R at 37°C for 1 h, and then the first strand of cDNA was synthesized by routine SuperScript First-Strand Synthesis System (Takara Biotechnology) for RT-PCR Kit for circRNA amplification. The real-time quantitative PCR was performed with SYBR-Green Premix Ex Taq (Takara Biotechnology Co., Ltd) and the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad Laboratories) according to the manufacturer's protocol. The PCR primers for hsa_circ_0001173 were AGAGGCTACGGGAGGAGAAC (forward) and TCTGCTGGCAATTCAAACAC (reverse). The primers for hsa_circ_0007291 were TCATGTCTGTGGGACTCTGC (forward) and CCAGGGAAACACTGAGGACT (reverse). The primers for hsa_circ_0003550 were ACGGGAAGCGATGATAACTG (forward) and TGGTATCCCAGATCCTCAGC (reverse). The primers for hsa_circ_0001947 were CAGTGCAGCCAGTTCAAAGA (forward) and AGTTTCCAAGCGTGTTCTGG (reverse). The primers for the ß-actin gene were GGCGGCACCACCATGTACCCT (forward) and AGGGGCCGGACTCGTCATACT (reverse). The PCR reaction required 10 μl 2 × SYBR-Green Premix EX Taq reagent, 50 ng DNA, 1 μl of gene primer in a total volume of 20 μl. The reaction conditions included initial denaturation at 95°C for 30 s, 40 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 30 s, followed by 81 cycles of dissociation at 55°C for 30 s. The specificity of amplification was determined by analyzing the dissolution curve of the product. The relative expression of circRNA was calculated by CT value method, and each experiment was repeated at least three times.
Target prediction of circRNAs
The circRNA–miRNA–mRNA network was developed based on the same expression trend between circRNA and its corresponding mRNA. The circRNAs interacted miRNAs were predicted using cytoscape.11 The regulatory network between miRNA and its downstream genes was predicted by mirdb software.12 Meanwhile, interactive scores were provided to evaluate accuracy, and only differential expression genes (DEGs) with scores greater than 90 were selected for further analysis.
Functional enrichment analysis and survival analysis
Based on the DEGs in the preliminary circRNA–miRNA–mRNA regulatory network, a further analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was performed using ggplot2 software.13 In addition, to evaluate the prognostic value of the circRNAs, the influence of circRNAs parental genes on survival of thymoma patients was also analyzed. The TCGA thymoma (PRAD) data was selected in UCSC Xena14 to obtain the Kaplan–Meier plot of the circRNA parental gene.
Statistical analysis
All the data were analyzed using the Statistical Product and Service Solutions (SPSS) 20.0 software package (IBM SPSS) and GraphPad Prism 6.0 (GraphPad Software, Inc.). All data were expressed as the mean ± SD. A Student's t-test was used to compare the measurement data between the two groups. The threshold of significance was defined as a p-value <0.05.
RESULTS
Differential expression profile of circRNA in thymus tissue of patients with thymoma
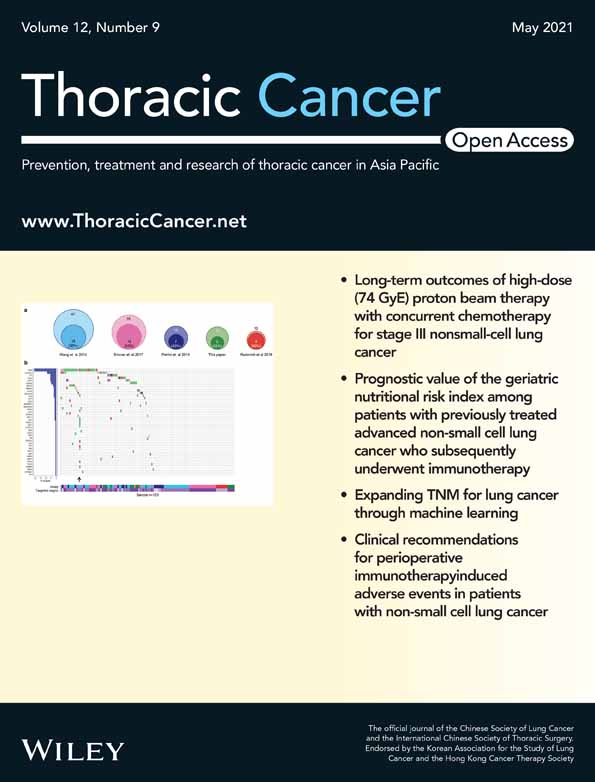
In total, 73 circRNAs were differentially expressed in both thymoma patients, of which 35 were upregulated and 38 were downregulated (Figure 1(a)). Since the upregulated circRNAs are more suitable as biomarkers than the downregulated circRNAs, the top four circRNAs from upregulated circRNAs were selected for further analysis (Table 1). Figure 1(b) demonstrated that the expression levels of four selected circRNAs were significantly upregulated in thymoma tissue, but there was no significant difference in the corresponding parental genes. Next, we investigated the exon composition of these four circRNAs. Most circular RNAs contained multiple exons, only hsa_circ_0001947 contained one annotated exon (Figure 1(c)). Similarly, the heat map results showed the expression levels of selected four circRNAs in thymoma tissue were higher than that in normal tissues in both thymoma patients (Figure 1(d)).
| circRNA ID | Chrom | Start | End | Strand | Cognate mRNA | Parental gene |
|---|---|---|---|---|---|---|
| hsa_circ_0001173 | chr20 | 58 438 944 | 58 441 083 | + | ENST00000475243.5 | VAPB |
| hsa_circ_0007291 | chr3 | 47 425 484 | 47 428 670 | − | ENST00000265565.10 | SCAP |
| hsa_circ_0003550 | chr4 | 10 097 710 | 10 103 986 | − | ENST00000499869.7 | WDR1 |
| hsa_circ_0001947 | chrX | 148 661 907 | 148 662 768 | + | ENST00000370460.7 | AFF2 |
Validation of the differentially expressed circRNAs
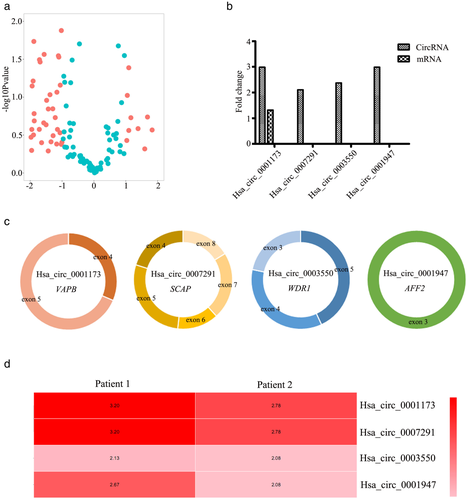
The potential candidate circRNAs from high-throughput sequencing were validated using specific divergent primers spanning the back-splice junction site.15, 16 As shown in Figure 2, the expression levels of hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 were significantly upregulated in thymoma tissue compared with control tissues (p-values were all less than 0.01).
Prediction of the circRNA-miRNA-mRNA interaction
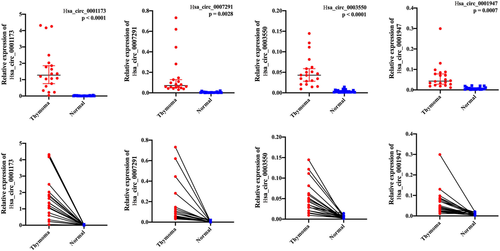
CircRNA can bind to the corresponding microRNA like a sponge and participates in the regulation of target gene expression, which is called the competitive endogenous RNA (ceRNA) mechanism. To further investigate the functions of selected circRNAs, the present study investigated the potential miRNAs binding with the circRNA based on the miRNA response element. As shown in Figure 3, a total of four circRNAs and 214 miRNAs were involved in the network, of which most of the miRNAs were regulated by hsa_circ_0007291 and hsa_circ_0001947 according to the p-value. At the same time, computational miRNA target mRNA analysis was performed by combining mRNA and circRNA expression profiling. In total we obtained 3456 possible miRNA-mRNA target pairs with the mirdb software (mRNAs are not shown in the figure due to the large number). Some mRNAs were associated with more than one miRNA, such as NM_001455 was targeted by hsa-miR-1-5p, hsa-miR-182-5p, hsa-miR-21-3p and hsa-miR-5006-5p.
Pathway analysis of differentially expressed genes
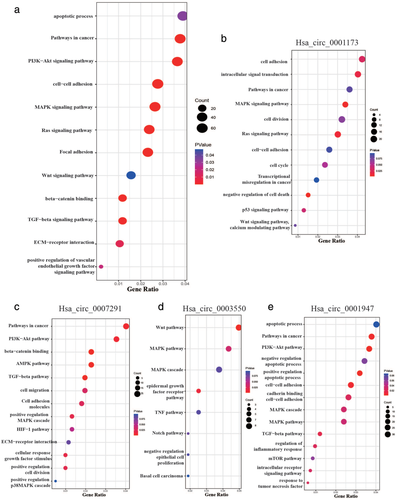
In order to further explore the potential pathogenesis of thymoma, we analyzed the biological function of 1792 DEGs in the regulatory network through KEGG pathway analysis. KEGG analysis demonstrated that there were 475 enrichment items from all four circRNAs that were statistically significant (p < 0.05), including the apoptotic process, pathway in cancer, PI3K-AKT signal pathway, and so on (Figure 4(a)). Meanwhile, a total of 172, 264, 79, and 280 enrichment items were involved in the downstream pathway of hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947, respectively. The top KEGG pathway items that were the most meaningful are shown in Figure 4(b)–(e). Among them, cell–cell adhesion, MAPK pathway, TNF pathway etc. attracted our attention because of their participation in immune response.
Survival analysis of parental genes of circRNAs
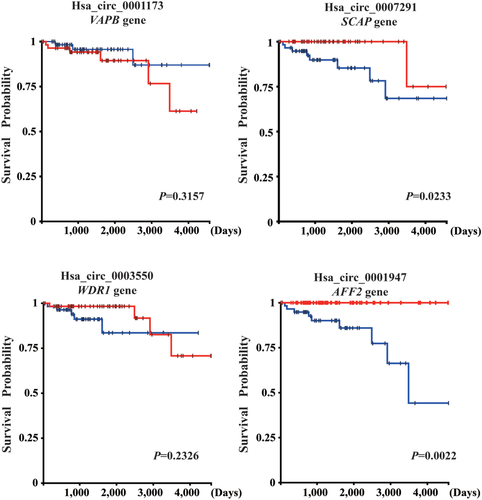
To further explore the function of circRNA, we examined the survival rates of VAPB, SCAP, WDR1, and AFF2, the parental genes of hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947. As shown in Figure 5, SCAP and AFF2 were significantly associated with overall survival using the Kaplan–Meier survival analysis. Patients with high expression of SCAP and AFF2 showed better overall survival. The results indicated that circRNA might be involved in the pathogenesis of thymoma by transcriptionally regulating the expression of parental genes or some other way. However, this hypothesis needs to be confirmed by further studies.
DISCUSSION
The present study demonstrated that hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 were expressed at high levels and associated with important pathological pathways in thymoma patients. Furthermore, the expression of the parental gene of hsa_circ_0007291 and hsa_circ_0001947 was associated with survival rate of thymoma patients. Hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 may not only be a potential biomarker, but also may be involved in the pathogenesis.
The thymus is where T cells differentiate, develop and mature, and it also secretes thymic hormones with immunomodulatory function, thus it is closely related to immunity. In the pathway analysis of circRNA regulatory genes, we focused on immune-related signaling pathways. Among them, cell–cell adhesion, MAPK pathway and tumor necrosis factor (TNF) pathway were of great interest because of their participation in immune response. Cell adhesion is a critical step in many immunological processes, including the binding of leukocytes to endothelial cells, infiltration of cells into inflammatory tissues and cytotoxicity to target cells.17 MAPK activation induces the expression of multiple genes that together regulate the inflammatory response.18 TNF plays an important role in the typical immune response by regulating the immediate inflammatory response involved in a series of significant innate immunity, as well as the subsequent activation of proliferation and programmed cell death or necrosis.19 Therefore, it is very likely that hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 participate in the abnormal immune regulation of thymus through these signal pathways.
Previous studies have revealed that circRNA performs its biological function mainly through the following three aspects. First, circRNA cis-regulates the expression of its parental gene20, 21 second, circRNA adsorbs miRNA through the mechanism of ceRNA22, 23 and third, CircRNA can also regulate gene transcription by binding to RNA binding proteins.24, 25 Although the differential expressions of circRNAs were obvious in thymoma tissues and normal tissues, the mRNA expression levels of their parental genes were not obvious (Figure 1(b)), indicating that the cis-regulation of these four circRNAs on their parent genes was not strong. Therefore, we speculated that these four circRNA function mainly through their downstream miRNAs-mRNA pathway.
At the same time, we found that SCAP (hsa_circ_0007291 parental gene) and AFF2 (hsa_circ_0001947 parental gene) were all significantly related with progression-free survival (PFS) of thymoma patients in the Kaplan-Meier plot. SCAP has been shown to specifically regulate the innate immune signal pathway induced by cytosolic cicrobial DNAs stimulation26, 27 and protect mice from systemic inflammation.28 The DEK–AFF2 fusion was the likely driver event in the squamous cell carcinoma of head and neck29 and stimulated T cell activation by interferon-γ (IFN-γ) secretion.30 Therefore, hsa_circ_0007291 and hsa_circ_0001947 may also participate in immune disorders by affecting the instability of the parental gene SCAP and AFF2.
In the preliminary exploration of this study, hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 in thymoma tissues showed some potential as biomarkers of disease, such as high specific expression, sensitive detection, and correlated with patient survival rates. However, the number of samples used in this study was small, which will affect the accuracy of the analysis results. The present study results need to be further verified in larger cohorts. Moreover, in addition to the prediction of bioinformatics software, further experiments are needed to confirm the relationship between CircRNAs and miRNAs. Whether hsa_circ_0001173, hsa_circ_0007291, hsa_circ_0003550, and hsa_circ_0001947 can be used as biomarkers of thymoma patients in clinical diagnosis and treatment remains to be further studied.
In summary, elucidating the specific molecular mechanism of immune response from the perspective of circRNA can provide a more effective diagnosis, prevention and treatment for thymoma, and it is also of great value to the research and development of drugs to improve thymoma therapy.
ACKNOWLEDGMENTS
This work was supported in part by grants BJ-2018-137 and BJ-2019-149 from the Beijing Hospital Nova Project, grant 2019WJGW-10-01 from the Beijing Dongcheng District Outstanding Talent Funding Project, grants 81871107 and 81870048 from the National Natural Science Foundation of China, and grant 2017ZX09304026 from the National Science and Technology Major Project for Significant New Drugs Creation, grant 2018-I2M-1-002 from the CAMS Innovation Fund for Medical Sciences.
CONFLICT OF INTEREST
The authors had no conflicts of interest and no disclosures to declare.