Tumor-educated leukocytes mRNA as a diagnostic biomarker for non-small cell lung cancer
Limin Niu and Wei Guo contributed equally to this work.
Funding information: National Natural Science Foundation of China, Grant/Award Number: 81773237; Jinan Science and Technology Program, Grant/Award Number: 201704080; the Medicine and Health Science Technology Development Program of Shandong Province, Grant/Award Number: 2017WS001
Abstract
Background
This study aimed to investigate the diagnostic and prognostic role of tumor-educated leukocytes (TELs) mRNA in Chinese patients with non-small cell lung cancer (NSCLC).
Methods
The TELs collected underwent total RNA isolation. RNA-sequencing (RNA-seq) technology was used to analyze the transcriptome of the TELs. The mRNA expression levels of differential genes were analyzed by RT-qPCR. Statistical analyses were performed using Prism and SPSS by Mann–Whitney nonparametric test, Kruskal-Wallis test and one-way ANOVA.
Results
We used RNA-seq technology to screen 95 differential genes (DEGs) from seven NSCLC and four controls, wherein 15 genes were upregulated, and 80 were downregulated. Of these, four genes were selected for further analysis, wherein one was upregulated (GPX1) and three were downregulated (BCL9L, MAP3K7CL, PCSK7). RT-qPCR was performed in 431 samples (237 NSCLC, 194 healthy donors). The four-gene panel showed significant differences (p < 0.001) in the expression levels between NSCLC and healthy samples. ROC curves of the panel revealed an AUC of 0.803, with a sensitivity of 73.8% and specificity of 75.3%. GPX1, BCL9L and PCSK7 genes distinguished early-stage NSCLC patients from healthy group (p < 0.05). When the three genes were combined to diagnose early-stage NSCLC, the diagnostic efficacy was 0.772, sensitivity was 73.7%, and specificity was 72.2%. In addition, the downregulated gene BCL9L was associated with chemotherapeutic effect.
Conclusions
The present study provided a systematic description of gene expression profiling in the TELs. It is worth noting that these four genes may be potential candidate genes for NSCLC diagnostic biomarkers and provide a basis for further biological and functional studies.
INTRODUCTION
According to the annual report on the 2020 Cancer Statistics by Siegel et al., lung cancer ranks first among male and female cancer deaths.1 Lung cancer includes non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), of which NSCLC accounts for about 80%.2 Due to the high degree of malignancy and rapid progression of lung cancer, almost two-thirds of patients are diagnosed at an advanced stage (stage III/IV). Despite continuous improvements in treatment strategies such as surgery, chemotherapy, radiotherapy and targeted therapy, the prognosis for patients diagnosed with NSCLC is extremely poor, with a five-year survival rate of less than 15%.3 Detection of lung cancer at an early stage can reduce mortality by 10 to 50 times,4, 5 which is important for the prevention, treatment, prognosis and improvement of patients' quality of life, and is the best way to improve the five-year survival rate of lung cancer. Thus, a sensitive, specific, blood-based, noninvasive test for the early detection and prediction of prognosis may assist medical oncologists in diagnosing the disease at an early stage, or in providing more aggressive treatment to effectively control the disease.
Leukocytes, derived from bone marrow hematopoietic stem cells, are a very important class of blood cell in human blood, which include neutrophils, eosinophils, basophils, monocytes, B cells, and T cells. Leukocytes are considered the main components, constituting the first line of defense in the immune system. These nucleated cells contain a considerable number of genes encoded in the human genome with detectable levels of transcripts and are easy to use in screening processes. At present, many studies have shown that there are significant differences in gene expression profiles between normal and malignant tissues of patients with primary tumors, and tumor tissue gene expression profiles are associated with overall patient survival.6 Peripheral blood is rich in leukocytes, which first causes changes in leukocytes gene expression profiles during the body's immune editing process. Therefore, it may be meaningful to replace the expression profiles in tissues for the diagnosis or prediction of tumor patients. Several studies have been reported on transcriptome analysis using peripheral blood cells in disease prediction and cancer classification. The expression levels of many peripheral blood mononuclear cell (PBMC) transcripts have the capability of acting as predictors for the outcome of time to progression and overall survival in renal cancer patients, whereas the prognostic value of peripheral blood expression profiles has also previously been verified in both leukemia and lymphoma.7, 8 In addition, alterations in the expression profiles of leukocytes have previously been observed in a wide range of cancers, including bladder9 and colorectal cancers.10 During immune editing, leukocytes are educated by tumor cells, and their RNA expression profiles change significantly, and they are known as tumor-educated leukocytes (TELs). At the same time, thanks to its closed membrane structure, the biological information related to tumors can be completely preserved. For these reasons, the substances carried by leukocytes have great potential to become a new tumor biomarker.
RNA-sequencing (RNA-seq) can accurately quantify the expression levels of genes and establish a global view of the whole genome.11 It has become a powerful tool to crack the global pattern of gene expression, including the discovery of an unprecedented global view of the transcriptome, expression and sequence variation of allele-specific expression.12-14 RNA-seq not only assists in mapping and transcriptome annotation, but also in understanding many biological processes, and is widely used to search for novel cancer biomarkers as well as monitor their development.
Hence, in this study, we aimed to set up a blood-based, disease-specific diagnostic screening method to accurately classify NSCLC patients from healthy donors. We selected four differential genes from leukocytes between NSCLC patients and healthy donors using RNA-seq, and then compared the expression patterns of these four differential genes in the leukocytes of NSCLC patients with various clinical variables to determine whether the mRNA expression patterns were correlated with eventual patient outcome.
METHODS
Patients and healthy donors
A total of 244 patients with NSCLC and 198 healthy donors at the Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences from April 2018 to December 2020 were enrolled. Patients with NSCLC were confirmed by combined clinical, pathological, and radiological diagnostic approaches, and the tumor stage was determined according to the eighth edition of the lung cancer TNM staging standards formulated by IASLC. Patients did not undergo any anticancer treatment or have any other endocrine, immune, or metabolic diseases. The healthy donors did not present with any other disease. Informed consent was obtained from all individuals. The demographic characteristics of included patients and healthy donors are described in Table 1.
| No. (%) of patients and healthy participants | |
|---|---|
| NSCLC | 237 |
| Male | 145 (61.2) |
| Age, median (min, max) | 61 (34, 86) |
| Behavioral factors | |
| Smoking | 113 (47.7) |
| Non-smoking | 124 (52.3) |
| T category | |
| T1 | 81 (34.2) |
| T2 | 62 (26.2) |
| T3 | 23 (9.7) |
| T4 | 50 (21.1) |
| Tx | 21 (8.9) |
| Extracapsular spread (for N1–N3) | |
| No | 71 (30) |
| Yes | 162 (68.4) |
| Nx | 4 (1.6) |
| Pathological type | |
| AC | 163 (68.8) |
| SCC | 51 (21.5) |
| Unknown | 23 (9.7) |
| Disease stage | |
| I | 57 (24.1) |
| II | 12 (5.1) |
| III | 63 (26.6) |
| IV | 105 (44.3) |
| Healthy participants | 194 |
| Male | 105 (54.1) |
| Age, median (min, max) | 51 (22,74) |
- Abbreviations: AC, adenocarcinoma; SCC, squamous cell carcinoma.
Blood collection and RNA isolation
Samples of peripheral whole blood were collected in EDTA vacutainer tubes and stored at 4°C until processing (within 6 h). RBC lysis buffer (NH4Cl, KHCO3, EDTA, PH = 7.2) was added at 3:1 volume ratio to lyse the RBCs. The samples were centrifuged at 450 g for 10 min at 4°C to yield a pellet containing leukocytes, and the pellet was washed several times with phosphate buffer. Total RNA was extracted from the leukocytes using Trizol reagent according to the manufacturer's protocol.
RNA preparation, library construction and illumina sequencing
A total amount of 2 μg RNA per sample was used for RNA sample preparations. Sequencing libraries were generated using NEBNext Mltra RNA Library Prep Kit for Illumina (NEB) according to the manufacturer's recommendations and the index codes were added to attribute sequences to each sample. Briefly, the mRNA was purified from the total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperatures in NEB Next First Strand Synthesis Reaction Buffer (5×). First strand cDNA was synthesized using random hexamer primer and RNase H. Second strand cDNA synthesis was subsequently performed using buffer, dNTPs, DNA polymerase I and RNase H. The cDNAs were assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies) and ABI StepOnePlus Real-Time PCR System (ABI). The libraries were sequenced on an Illumina Hiseq 4000 platform, and 150 base pair (bp) paired-end reads were generated.
Reverse transcription quantitative real-time PCR
A total of 1000 ng of RNA was reversed transcribed into cDNA using the Takara PrimeScript RT reagent Kit (Perfect Real Time, Takara) in 20-μl reaction. For the validation cohort, 2-ng cDNA was mixed with SYBR Green master mix (SYBR Green PCR Kit, Takara) and primers in a 20-μl reaction volume. PCR amplification was done on LightCycler 480 QPCR system (Roche Diagnostics). Dissociation curves generated at the end of each cycle were examined to verify specific PCR amplification and absence of primer-dimer formation. The sequences of the four primer sets are listed in Table S1. Total RNA isolated from each sample was run in two replicates, and then the median Ct was calculated. The transcription level of ACTB was used as reference genes. The relative expression was calculated using the comparative cycle threshold (ΔCt) method.
Data reporting and statistical methods
The GraphPad Prism version 6.0 (GraphPad Software) and SPSS 22.0 software (IBM) were used for statistical analysis. Data are shown as medians with interquartile range. Mann–Whitney test was used to compare between the two groups. For comparisons between more than two groups, nonparametric Kruskal-Wallis test or the parametric one-way analysis of variance (ANOVA) test were used. Receiving operating characteristic (ROC) curve analysis was then used to evaluate the discriminatory power of the combinations. p values < 0.05 were considered to be statistically significant and all tests were two-sided.
RESULTS
Identification of differentially expressed genes using RNA sequencing
To screen differential genes, leukocytes were collected after erythrocyte lysis from seven NSCLC patients and four healthy donors, respectively, and were subjected to RNA sequencing. The demographic characteristics of seven patients are described in Table S2. The expression of each gene was measured by fragments per kilobase of transcript per million mapped reads (FPKM). An average of 34 018 genes were identified. A total of 95 genes were differentially expressed between the control and cancer samples, wherein 15 genes were upregulated and 80 were downregulated in the NSCLC group (|log2Foldchange| >0 and p < 0.01) (Figure 1(a)). The expression patterns of these 95 differentially expressed genes (DEGs) are shown as a heatmap using hierarchical cluster analysis (Figure 1(b)).
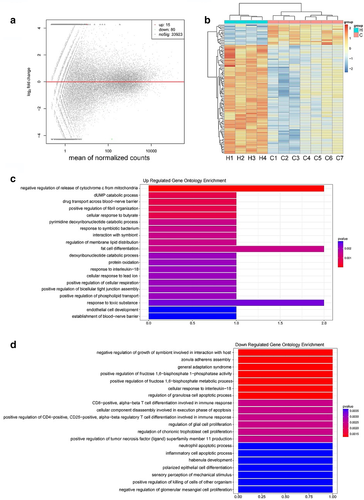
Next, the gene ontology (GO) functional classification was performed to gain overall insight into the function of annotation genes. A total of 152 clusters were significantly annotated with GO function of biological process (BP) (p < 0.01). The results of GO analysis demonstrated that the upregulated enrichment was enriched in specific biological processes including the negative regulation of release of cytochrome c from mitochondria, fat cell differentiation, response to toxic substances (Figure 1(c)), and downregulation with CD8-positive, αβT cell differentiation involved in immune response and cellular component disassembly involved in the execution phase of apoptosis (p < 0.01) (Figure 1(d)).
Validation of differentially expressed genes in NSCLC and healthy donor groups using RT-qPCR
The selected mRNA expression levels were then analyzed by RT-qPCR. Of the 95 DEGs, some genes according to the expression differences with greater than two folds between the groups were selected. Then four genes (glutathioneperoxidase1 [GPX1], B-cell CLL/lymphoma 9-like [BCL9L], MAP3K7 C-terminal like [MAP3K7CL], and proprotein convertase subtilisin/kexin type 7 [(PCSK7]) were selected due to a more significant difference in their expression. GO analysis demonstrated that the four genes were enriched in biological processes including regulation of apoptotic process, and cellular response to oxidative stress.
Next, these four genes were validated in an expanded cohort with 237 NSCLC patients, including 57 patients with early-stage NSCLC (stage I) and 194 healthy donors. The relationship between mRNA expression and the clinical factors is shown in Table 2. The expression of GPX1, MAP3K7CL and PCSK7 in the TELs showed no relationship with age, gender, smoking history, pathological type and metastasis (p > 0.05). Nevertheless, the expression of BCL9L was related to gender (p = 0.0001) and progression of the disease (p = 0.043) but showed no relationship with other clinical factors.
| n | p-value | ||||
|---|---|---|---|---|---|
| GPX1 | BCL9L | MAP3K7CL | PCSK7 | ||
| Age | 0.221 | 0.072 | 0.400 | 0.403 | |
| ≤61 | 116 | ||||
| >61 | 121 | ||||
| Gender | 0.534 | 0.0001 | 0.269 | 0.079 | |
| Male | 145 | ||||
| Female | 92 | ||||
| Smoking history | 0.175 | 0.032 | 0.867 | 0.506 | |
| Smoker | 113 | ||||
| Non-smoker | 124 | ||||
| T category | 0.434 | 0.005 | 0.015 | 0.069 | |
| T1 | 81 | ||||
| T2 | 62 | ||||
| T3 | 23 | ||||
| T4 | 50 | ||||
| Nodal status | 0.326 | 0.085 | 0.118 | 0.556 | |
| N0 | 71 | ||||
| N1 | 162 | ||||
| Distant metastasis | 0.537 | 0.383 | 0.671 | 0.360 | |
| M0 | 132 | ||||
| M1 | 105 | ||||
| Disease stage | 0.284 | 0.043 | 0.092 | 0.496 | |
| I | 57 | ||||
| IIa–IV | 180 | ||||
| Pathological type | 0.403 | 0.065 | 0.240 | 0.058 | |
| AC | 163 | ||||
| SCC | 51 | ||||
- Abbreviations: AC, adenocarcinoma; SCC, squamous cell carcinoma.
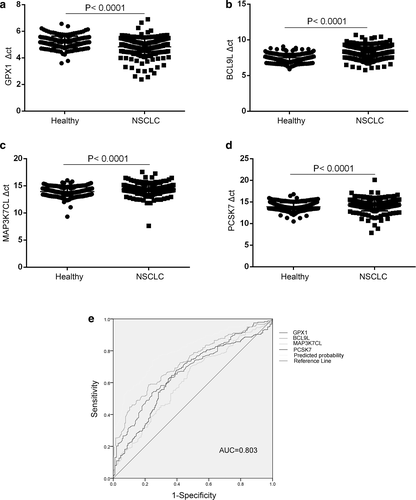
Following Mann–Whitney tests, the expression of GPX1 genes were upregulated in NSCLC, showing significant differences when cancer patients were compared with healthy groups (Figure 2(a), p < 0.0001), whereas the expression of BCL9L, PCSK7 and MAP3K7CL genes were downregulated in NSCLC, showing significant differences (all p < 0.0001) between the two cohorts (Figure 2(b–d)). To evaluate the diagnostic performance of the combination panel of differential mRNAs for NSCLC, a ROC curve was calculated by comparing the 237 NSCLC samples with 194 healthy donor samples. As shown in Figure 2(e), the AUC was 0.803, with a sensitivity of 73.8% and specificity of 75.3%. Taken together, these data suggested that the expression levels of these four genes might be different, and that it might therefore be possible to distinguish NSCLC patients from healthy donors.
mRNA expression levels of TELs associated with tumor stage
Next, we analyzed the different expression levels of these four genes in early stage NSCLC patients and compared them with those in healthy donors to explore their diagnostic role in NSCLC. A total of 194 healthy donors and 57 early-stage NSCLC patients were grouped and subjected to pooled analysis. As shown in Figure 3(a–c), GPX1 genes were highly expressed in the early-stage NSCLC patients, whereas BCL9L and PCSK7 was expressed at low levels compared with that in healthy donors (p < 0.0001, p < 0.0001, and p = 0.0065, respectively). When the three genes were combined to diagnose early-stage NSCLC, the diagnostic efficacy was 0.772, sensitivity was 73.7%, and specificity was 72.2% (Figure 3(h)), indicating a potential diagnostic role of these three genes in early-stage NSCLC. Unexpectedly, MAP3K7CL gene expressions in these two groups showed no statistical significance (data not shown).
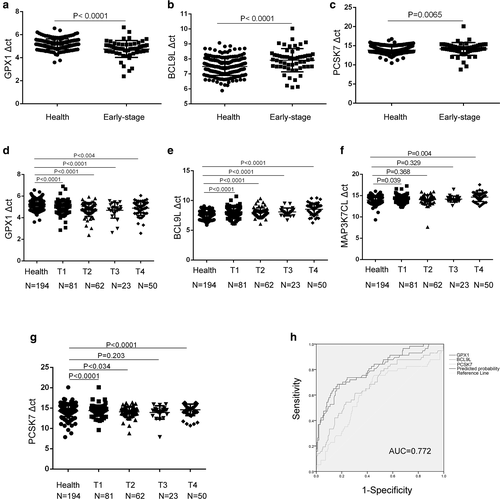
Moreover, we further analyzed the relationship of the expression of these four genes with T stages. Our data showed that GPX1, BCL9L gene expression levels demonstrated statistical significance in T1–T4 NSCLC patients when compared to healthy donors (Figure 3(d,e)). MAPK7CL and PCSK7 expression was significantly lower in T1, T4 NSCLC patients than in healthy donors (Figure 3(f,g)). These results indicated that the mRNA levels of white blood cells might be used in the early detection of tumors.
Role of BCL9L in the prediction of response to chemotherapy
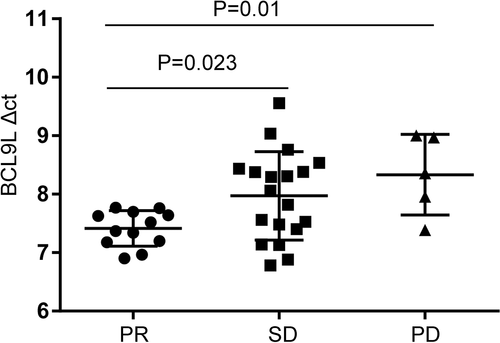
In our study, 36 patients with NSCLC received first-line chemotherapy. Clinical response criteria were defined according to the RECIST guidelines. Partial response (PR) was defined as reduction in the size of all measurable tumor areas by at least 30%, progressive disease (PD) was defined as an increase of ≥25%, compared to baseline or best response in the size of all measurable tumor areas, and stable disease (SD) was defined as neither adequate shrinkage to qualify for PR nor adequate increase to qualify for PD. Five patients showed no response to first-line chemotherapy PD, 19 patients had SD and 12 patients had a PR. The results suggested that only the BCL9L gene was associated with chemotherapeutic effect, and that the BCL9L expression levels were lower in the PR group than that in the SD and PD groups (p = 0.023, p = 0.01) (Figure 4). These data indicated that BCL9L might act as a potential biomarker in the prediction of a response to chemotherapy.
DISCUSSION
Despite continuous improvements in the treatment of NSCLC, patients remain extremely vulnerable to relapse and mortality. Thus, sensitive and specific biomarkers to identify NSCLC in patients are urgently needed. In the current study, we identified and validated several DEGs in the leukocytes between the healthy donors group and NSCLC patients. We reported that these four genes were differentiated in the two groups with an AUC of 0.803. This four-gene set represents a novel biomarker in the detection of NSCLC.
Leukocyte mRNAs acted as a biomarker for the early diagnosis of NSCLC with obvious advantages. The occurrence and development of tumors are usually accompanied by an immune response, as well as the continuous interaction between white blood cells and tumor cells, triggering subtle changes in gene expression in white blood cells. Thus, their specific expression patterns are capable of reflecting, and even enriching, the static and dynamic changes in tumors due to their high abundance in circulation.15 These advantages make the leukocyte mRNAs useful for diagnostic purposes. Therefore, our research provided a novel blood-based test for NSCLC.
In our study, we used RNA-seq technology to screen the DEGs in the leukocytes. To our knowledge, this is the only study which has provided a systematic description of the global transcriptome of differential genes in the leukocytes between healthy subjects and NSCLC patients. We found that a total of 95 genes were differentially expressed in the leukocytes between control and NSCLC samples, wherein 15 genes were upregulated and 80 were downregulated in NSCLC patients. These findings indicate that the initial data was successfully obtained with RNA-seq technology, which has laid a solid foundation for subsequent experiments.
Next, four differential mRNAs from the blood cells were verified by RT-qPCR, including GPX1, MAP3K7CL, BCL9L and PCSK7. Previous studies have already confirmed the role of these genes in cancer. Human cellular GPX1 is an important member of the natural enzymatic antioxidants which protect the cells from oxidative damage and may be involved in cell signaling and reducing inflammatory processes.16-18 There have been very few research studies that have demonstrated the relationship between GPX1 and NSCLC. Our study showed a significant difference between the healthy donor and NSCLC groups, indicating that GPX1 might be the NSCLC response gene. However, it was unclear as to how GPX1 took part in the tumor progression. PCSK7, a member of the subtilisin-like proprotein convertase family that processes multiple protein precursors,19 is expressed in the organs that are largely involved in lipid metabolism, such as colon, lymphoid-associated tissues, liver and the intestine.20 Currently, there are no research studies which discuss the relationship between PCSK7 and any other tumor. Our data demonstrated that the expression of PCSK7 was lower in NSCLC patients compared to the healthy donor group, and might provide a new breakthrough in tumorigenesis. BCL9L is a nuclear WNT pathway component that enhances proliferation, survival, migration, invasion, and metastatic potential of tumor cells. It has been reported to be upregulated in oral squamous cell carcinoma,21 multiple myeloma and colon carcinoma primary tumors.21, 22 However, in current study BCL9L expression was downregulated in leukocytes of NSCLC patients , implying its different expression between in tumor tiusse and in leukocyte. It was conceivable that these four genes in the leukocytes may contribute to the pathogenesis of NSCLC via reducing inflammatory processes, regulating mitochondrial ROS signaling and mediating the WNT pathway. Our finding suggested that white blood cells have unique characteristics that potentially make them a new diagnostic tool.
Interestingly, we found that the BCL9L gene was associated with chemotherapeutic effect. BCL9L is required for efficient β-catenin-mediated transcription in human cell lines whose Wnt pathway remain active.23 The Wnt/β-catenin pathway is an important pathway in cancer, and there are no well-established, validated small molecule inhibitors of this pathway to date. The BCL9L gene has been reported to be upregulated in a variety of human cancers, highlighting its importance as a novel therapeutic target in the treatment of cancers associated with deregulated Wnt signaling. Low expression of BCL9L in NSCLC after first-line drug therapy showed the increased possibility of tumor recurrence. Further validation of the BCL9L gene as a drug target in NSCLC is warranted, including the analysis of their function in Wnt signaling and its role in promoting tumorigenesis. The functional importance and lower expression of BCL9L in NSCLC indicated it as a potential biomarker to predict chemotherapeutic effect and may be a potential gene as a cancer drug target.
In summary, in this study we identified a peripheral blood biomarker panel in order to differentiate NSCLC patients from healthy donors. Further confirmatory studies are required as to whether this blood-based test will be valuable for screening candidates for NSCLC. The increased rates of compliance expected for a blood test when compared to other cancer screening modalities potentially results in the early detection of cancer, decreasing the morbidity and mortality and more effectively utilizing the health care resources. Further work should be carried out to refine the algorithm for choosing optimal marker combinations to be incorporated into a NSCLC biomarker panel. The manipulation of leukocytes may represent a potential therapeutic strategy that deserves further investigation.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81773237), the Medicine and Health Science Technology Development Program of Shandong Province (2017WS001), and Jinan Science and Technology Program (201704080).
CONFLICT OF INTEREST
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.