Chromosome-level genome assembly of the aphid parasitoid Aphidius gifuensis using Oxford Nanopore sequencing and Hi-C technology
ABSTRACT
Aphidius gifuensis is a parasitoid wasp that has been commercially bred and released in large scale as a biocontrol agent for the management of aphid pests. As a highly efficient endoparasitoid, it is also an important model for exploring mechanisms of parasitism. Currently, artificially bred populations of this wasp are facing rapid decline with undetermined cause, and mechanisms underlying its parasitoid strategy remain poorly understood. Exploring the mechanism behind its population decline and the host–parasitoid relationship is impeded partly due to the lack of a comprehensive genome data for this species. In this study, we constructed a high-quality reference genome of A. gifuensis using Oxford Nanopore sequencing and Hi-C (proximity ligation chromatin conformation capture) technology. The final genomic assembly was 156.9 Mb, with a contig N50 length of 3.93 Mb, the longest contig length of 10.4 Mb and 28.89% repetitive sequences. 99.8% of genome sequences were anchored onto six linkage groups. A total of 11,535 genes were predicted, of which 90.53% were functionally annotated. Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis showed the completeness of assembled genome is 98.3%. We found significantly expanded gene families involved in metabolic processes, transmembrane transport, cell signal communication and oxidoreductase activity, in particular ATP-binding cassette (ABC) transporter, Cytochrome P450 and venom proteins. The olfactory receptors (ORs) showed significant contraction, which may be associated with the decrease in host recognition. Our study provides a solid foundation for future studies on the molecular mechanisms of population decline as well as host–parasitoid relationship for parasitoid wasps.
1 INTRODUCTION
Parasitoid wasps have been widely used as effective biological control agents against numerous destructive pests in the agricultural industry, including species of Lepidoptera, Diptera, Coleoptera and Hemiptera (Beckage & Gelman, 2004; Wang et al., 2019). The parasitoid wasps lay eggs inside or on the surface of pests, and the parasitoid larvae absorb nutrients from their hosts until emergence. To increase the efficiency of parasitism, the wasps develop a variety of strategies, producing and transmitting bioactive substances into hosts, such as venom (Asgari & Rivers, 2011; Mrinalini, & Werren, 2015), polydnavirus (Belle et al., 2002), teratocytes (Dahlman et al., 2003; Strand, 2014), ovarian proteins (Luckhart & Webb, 1996) and larval secretions. Multiple parasitoid wasps have been bred successfully on a commercial scale and widely applied to control specific agricultural pests (Wang et al., 2019), such as Aphidius ervi (Dennis et al., 2020), Cotesia flavipes (Muirhead et al., 2012), Encarsia formosa (Hoddle et al., 1998) and Trichogramma galloi (Postali Parra & Coelho, 2019).
Aphidius gifuensis Ashmead (Hymenoptera: Braconidae) is a common endoparasitoid of many destructive aphid species (Figure 1), including Myzus persicae, Sitobion avenae and Schizaphis graminum (Pan & Liu, 2014). In China, this wasp has been widely bred and used as a biocontrol agent for more than four decades to control the green peach aphid, Myzus persicae, a severe pest on vegetables and tobaccos (Li et al., 2018; Yang et al., 2011). The life history, physiological characteristics, parasitic ability and breeding technology of A. gifuensis have been extensively studied to improve its captive breeding behaviour and aphid control efficiency (Khan et al., 2016; Li et al., 2013; Pan et al., 2018; Yang et al., 2009). For instance, using comparative transcriptome and proteome analysis, Zhang et al. (2018) identified genes associated with diapause behaviour. Several chemosensory receptors and candidate genes associated with environmental adaptation were also identified, including chemosensory: olfactory receptors (ORs), gustatory receptors (GRs) and ionotropic receptors (IRs); detoxification systems: cytochrome P450, peroxidase and glutathione S-transferase; and heat shock proteins (Fan et al., 2018; Gao et al., 2017; Kang et al., 2017, 2018). Nevertheless, in recent years, commercially bred A. gifuensis has shown rapid population decline and a significant decrease in aphid control efficiency, which can be reflected by a marked decrease in its viability, fecundity and capability of recognizing and parasiting its aphid host (Xie et al., 2020). These issues have severely compromised the efficiency of A. gifuensis as a biocontrol agent in the field. However, knowledge about the molecular mechanisms of parasitism and population degradation of this wasp remains limited, in part due to the absence of the whole-genome information of A. gifuensis.

Whole-genome information plays an important role in genomic studies of the parasitoid wasps. Genome data can provide important insights into molecular mechanisms behind physiology and parasitic biology of parasitoid wasps. The unique haplodiploid mating system of the Hymenoptera is also beneficial to whole-genome sequencing. To date, whole genomes have been obtained for more than 40 parasitoid wasp species (Branstetter et al., 2018), seven of which were braconid wasps (Burke et al., 2018; Geib et al., 2017; Tvedte et al., 2019; Yin et al., 2018), including Aphidius ervi (Dennis et al., 2020), Cotesia vestalis (Shi, Wang, et al., 2019), Diachasma alloeum (Tvedte et al., 2019), Fopius arisanus (Geib et al., 2017), Lysiphlebus fabarum (Dennis et al., 2020), Macrocentrus cingulum (Yin et al., 2018) and Microplitis demolitor (Burke et al., 2018). Most braconid genomes were sequenced primarily on Illumina platforms, which precluded the assembly of target genome into chromosome-level scaffolds. The application of single molecule real-time sequencing technology can improve the quality, particularly in terms of contiguity, of the genome assembly.
In this study, we report a high-quality genome assembly of A. gifuensis, the first Aphidiinae genome reported at chromosome level, using Nanopore sequencing and Hi-C (proximity ligation chromatin conformation capture) technology. Comparative analyses with available wasp genomes were conducted to understand the genomic evolution of A. gifuensis. This reference genome will provide a foundation for future studies on the molecular mechanism of host recognition, parasitizing behaviour and population degradation of A. gifuensis, key features of this important biocontrol agent.
2 MATERIALS AND METHODS
2.1 Sampling and genome sequencing
All A. gifuensis samples for genome sequencing were collected from a laboratory-bred population reared on Myzus persicae in the Biological Control for Tobacco Diseases and Insect Pests Engineering Research Center of China Tobacco, Yunnan, China. About 17.4 μg genomic DNA was extracted from a pooled sample of 800 adult males using the Blood & Cell Culture DNA Kit (Qiagen). DNA was quantified by 0.75% agarose gel electrophoresis, Nanodrop spectrophotometry (Thermo Fisher) and Qubit 3.0 fluorometry (Invitrogen).
Using the TruSeq Nano DNA HT Sample preparation Kit (Illumina), a paired-end (PE) library, with an insert size of 350 bp, was constructed for genomic survey and sequenced with PE 150 bp on an Hiseq X Ten platform (Illumina). Libraries with long fragments were prepared and sequenced on a GridION X5 sequencer (Nanopore) at GrandOmics. The quality of these libraries was measured using 0.75% agarose gel electrophoresis.
The Hi-C sequencing library was prepared following Belton et al. (2012) with minor modifications. In brief, the cells of approximately 200 male samples were fixed with 2% formaldehyde for crosslinking. The fixed tissue was frozen in liquid nitrogen and ground to a powder before re-suspending in nuclei isolation buffer, following the protocol descripted by Shi, Ma, et al. (2019). The cross-linked DNA was digested with DpnII restriction endonuclease and marked by biotin-14-dCTP to remove nonligated DNA fragments. The ligated DNA was then sheared to 300–600 bp followed by a standard Illumina library preparation protocol, described in Meyer and Kircher (2010). The library was sequenced on the Illumina NovaSeq platform (Illumina) with paired-end (PE) 150 bp reads.
2.2 Genome size estimation and assembly
Raw data of short reads from the Illumina platform were filtered using strict quality controls by fastp v0.12.6 (Chen et al., 2018), to remove duplications, reads containing adapters and low-quality reads (-q 5 -u 50). These were then used to estimate the genome size based on the K-mer distribution analysed by kmerfreq v2.0 (Marçais & Kingsford, 2011). To assess the degree of microbial contamination, we checked 20,000 randomly selected Illumina sequencing reads by searching against the online NCBI-NR database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). We applied an e-value threshold of <1e-5, and according to this criterion, no microbial contaminant sequences were found in the sequencing reads. For genomic contig assembly, we first used canu v1.3 (Koren et al., 2017) to correct and trim the long sequencing reads with default parameters. The trimmed reads were assembled by using the software smartdenovo v1.0 (https://github.com/ruanjue/smartdenovo, -k 19, -j 5000, -e dmo). To correct for errors of assembly, the short reads were mapped to the genome assembly using bwa-mem v0.7.12-r1039 (Li & Durbin, 2009) with default parameters, followed by three iterations of polishing with Illumina short reads using nextpolish v1.0 (genome_size =auto, -min_read_len 10k, -max_read_len 150k, -max_depth 60) (Hu et al., 2019). The assembled contigs were also assessed against NCBI-NR database using the same approach as described for raw sequence reads and were again found to contain no microbial sequences. We calculated genome and assembly statistics, such as GC content and contig N50, using a custom python script (Supplemental text).
To further scaffold the assembly into chromosome-scale linkage groups, we performed Hi-C analysis. The raw Hi-C data were primarily filtered using the default parameters in Hi-C-Pro (Burton et al., 2013). Then, all the cleaned reads were mapped to the draft assembly contigs using bowtie2 v2.3.2 with end-to-end alignment mode (--very-sensitive -L 30) (Langmead & Salzberg, 2012), after quality control with Fastp as described previously. The uniquely aligned paired reads were retained for analysis. Based on the matrix of valid interaction reads, the contigs were anchored to chromosomal linkage groups using lachesis (https://github.com/shendurelab/LACHESIS) (Burton et al., 2013). The completeness of the final draft genome was assessed using busco v3.0.1 (Benchmarking Universal Single-Copy Orthologs) at nucleotide level based on 1658 genes in the insecta_odb9 database (Simão et al., 2015).
2.3 Transcriptome sequencing
Total RNA was extracted from 75 adult males using TRIzol Universal (Tiangen) (Rio et al., 2010). The NanoDrop spectrophotometer (Thermo Fisher) and Agilent 2100 Bioanalyzer were used to evaluate the quality of extracted RNA (OD260/280 = 2.0–2.2, OD260/230 = 1.8–2.1, 28 s:18 s ≥ 1.5, RIN ≥ 8). A total of 25.54 μg RNA was eluted with nuclease-free water. The PacBio Iso-Seq protocol was followed for library preparation: total RNA was reverse transcribed into cDNA using the Clontech SMARTer PCR cDNA Synthesis Kit. The cDNA was amplified for library construction with an insert size between 0.5 and 6 kb after size-selection using the BluePippin (Sage). The sequencing was performed on one SMRT cell on the PacBio Sequel platform (Pacific Biosciences) in circular consensus sequencing (CCS) mode. Raw sequence data were filtered with the standard IsoSeq3 protocol before subsequent analysis, including circular consensus calling through ccs, clustering and polishing through isoseq3 (https://github.com/ylipacbio/IsoSeq3).
2.4 Genome structure and functional annotation
repeatmasker vision 1.331 (https://github.com/rmhubley/RepeatMasker, Smit et al., 2013–2015), a de novo repeat library builder by repeatmodeler version-open-1.0.11 (http://www.repeatmasker.org/RepeatModeler, Smit & Hubley, 2008–2015) and repbase (http://www.girinst.org/repbase) (Bao et al., 2015), was used to identify repetitive sequences and transposable elements. LTR_Finder v1.06 (-C -w 0) (Xu & Wang, 2007) and MITE-Hunter (-n 20 -P 0.2 -c 3) (Han & Wessler, 2010) were applied to predict specific repeats. The repeat sequences were masked before genome annotation.
Gene structure prediction was performed using three strategies: homology-based prediction, ab initio prediction and transcriptome-based prediction. For homology-based prediction, gemoma v1.6.1 (Keilwagen et al., 2016, 2019) was used with default parameters and the published genomes from GenBank, including two model insects (Bombyx mori and Drosophila melanogaster) (Adams et al., 2000; Xia et al., 2004) and five hymenopteran species (Apis mellifera, Microplitis demolitor, Fopius arisanus, Nasonia vitripennis and Ceratosolen solmsi) (Table S1). We used each of the seven species as reference for homology-based prediction. The prediction based on Apis mellifera generated the highest BUSCO values and was used for the final integration with EVidenceModeler (EVM). Augustus v3.3.1 (--gff3=on --hintsfile=hints.gff --extrinsicCfgFile=extrinsic.cfg --min_intron_len=30 --softmasking=1 --allow_hinted_splicesites=gcag,atac) (Stanke & Waack, 2003) was used to predict genes in ab initio prediction, training parameters with the transcriptome data of A. gifuensis. Combining TransDecoder (http://transdecoder.github.io) with default parameters and pasa v2.3.3 (-c alignAssembly.config -C -R -g genome.fasta -T -u trans.fasta -t trans.clean.fasta -f fl.acc --ALIGNERS gmap) (Haas et al., 2003), the intact open reading frame (ORF) and retained de novo gene prediction were identified to improve the accuracy of prediction by using transcriptome data. Finally, we integrated the results of these three strategies using evidencemodeler (EVM) v1.1.1 (--segmentSize 1000000 --overlapSize 100000) (Haas et al., 2008).
Gene functional annotation was performed based on homologue searches and the best match to the databases of KEGG (Kanehisaa & Goto, 2000; Ogata et al., 1999), KOG (Tatusov et al., 2001), Swiss-Prot (Bairocha & Apweiler, 2000) and NCBI-NR using blastp (E-value <1e-5). Next, GO (Gene Ontology) analysis was executed through interproscan v5.32–71.0 (Zdobnov & Apweiler, 2001) to identify protein domains. The information from different sources of functional annotation was combined for each gene in the final integration.
Noncoding RNA, including transfer RNA (tRNA), microRNA (miRNA), ribosome RNA (rRNA) and small nuclear RNA (snRNA), was also identified. We annotated rRNA, miRNA and snRNA by mapping against the Rfam database (Kalvari et al., 2018) using BLASTN, predicted tRNA using tRNAscan-SE v2.0 (Lowe & Eddy, 1997) and built models to predict rRNA and subunits using rnammer v1.2 (Lagesen et al., 2007).
2.5 Orthology, synteny and phylogeny
Orthologous and paralogous gene families were identified by orthomcl v2.0.9 (percentMatchCutoff=50 evalueExponentCutoff=−5) (Li et al., 2003). The protein sequences of 13 hymenopteran species (Table S1) were downloaded from NCBI genome database. We filtered out alternative splicing for each gene, with the longest transcript kept to represent the coding region. We aligned proteins between A. gifuensis and 13 other hymenopteran species (Table S1) using blastall v2.2.26 (-p blastp -m 8 -e 1e-5 -F F, E-value <= 1e-5).
Protein sequences of the identified single-copy genes were aligned by mafft v7.313 with L-INS-i algorithm (Katoh & Standley, 2013). raxml v8.2.10 (-m PROTGAMMAAUTO -p 12345 -T 8 -f b) (Stamatakis, 2014) was employed to reconstruct the phylogenetic tree with 100 bootstraps, and two Symphyta species (Orussus abietinus and Cephus cinctus) were used as outgroups. We used the MCMCTree (clock =2, RootAge = <3.37, model =7, BDparas =1 1 0, kappa_gamma =6 2, alpha_gamma =1 1, rgene_gamma =2 3.606, sigma2_gamma =1 1.03) from the paml v4.8 package to estimate divergence time (Yang, 2007), using divergence time calibrated from the TIMETREE database. The minimum and maximum divergence times between Apis mellifera and Solenopsis invicta were 107–184 million years ago (Mya); Apis mellifera and Ceratosolen solmsi were 158–247 Mya; Apis mellifera and Orussus abietinus were 188–273 Mya (Hedges et al., 2006).
To identify collinear gene blocks between A. gifuensis, Apis mellifera and Nasonia vitripennis, we exported coding sequences (CDs), searched syntenic genes and visualized the high-quality blocks using MCscan (Multiple Collinearity Scan Toolkit) in JCVI (https://github.com/tanghaibao/jcvi) with default parameters.
2.6 Gene family expansion and contraction analysis
To identify the gene family expansion and contraction, protein sequences for A. gifuensis and 13 hymenopteran species were obtained from GenBank and other databases (Table S1). café (Computational Analysis of Family Evolution) v4.2.1 (-p 0.05 -t 10 -r 10000) (Hahn et al., 2007; Han et al., 2013) was used to compare generated gene family clusters, with a birth and death rate model estimated over a phylogeny. For gene families exhibiting significant expansion and contraction with Viterbi p-values < .05 (De Bie et al., 2006) in A. gifuensis genome, KEGG pathway enrichment and GO analysis were executed by r package clusterProfiler (Yu et al., 2012).
2.7 Gene family annotation
We manually annotated four gene families of cytochrome P450 monooxygenase (P450s), ATP-binding cassette (ABC) transporter, venom proteins and olfactory receptors (ORs). A set of orthologous protein sequences of related species (such as Apis mellifera, Aphidius ervi, Fopius arisanus and Lysiphlebus fabarum) from NCBI GenBank and Hidden Markov models (HMMs) were used as references for gene identification. blast v2.10.0 (E-value <1e-5) and hmmer v3.3 were used to search the candidate genes in the A. gifuensis genome. Then, the results of BLAST and HMMER analyses were integrated through the bioinformatic pipeline BITACORA (full mode) (Vizueta et al., 2020). The annotated candidate genes sequences were aligned using mafft v7.471 with G-INS-I strategy (Katoh & Standley, 2013). We constructed the phylogenetic tree using iq-tree v1.6.11 (-m TEST -bb 1000 -alrt 1000).
3 RESULTS AND DISCUSSION
3.1 Genome sequencing and assembly
A total of 18.6 Gb clean reads were generated by the Illumina platform. The genome size is approximately 154.9 Mb based on K-mer frequency distribution analysis (Figure S1). For long-read genome sequencing, we obtained 10.9 Gb Nanopore reads after removing low-quality sequences, which corresponds to approximately 69.5-fold coverage of the A. gifuensis genome (Table 1). The mean and N50 length of filtered reads were 16.7 kb and 23 kb, respectively. Under the correction and trimming of Canu and contig assembly of Smartdenovo, a 156.9 Mb draft genome was generated by de novo assembly, consisting of 136 contigs with a contig N50 of 3.93 Mb and longest contig of 10.4 Mb (Table 2). The average GC content was 26.5%. The resulting genome was slightly larger than the estimated size but smaller than most published genomes of parasitoid wasps. Our assembly of A. gifuensis genome using long-read Nanopore sequencing obtained a larger contig N50 compared to the other seven Braconidae genomes previously assembled using combined Illumina and PacBio sequencing methods (Table 3; Burke et al., 2018), in which only Microplitis demolitor and Cotesia vestalis had a scaffold N50 longer than 1 Mb (Burke et al., 2018). The other five genomes had contig N50 s ranging from 192.4 to 980 kb. Our results also highlight the importance of using long-read sequencing technology which largely increased the quality of assembly compared with solely using Illumina sequencing. Along with the other two Aphidiinae species, Aphidius ervi and Lysiphlebus fabarum, the genomic GC content was also particularly low (<27%). There may be biological factors which further contribute to the low GC content. For some bacteria (Almpanis et al., 2018; Barahimipour et al., 2015; McCutcheon et al., 2009) and plant species (Šmarda et al., 2014; Veleba et al., 2017), environmental conditions, such as limited nitrogen availability, could contribute to the low GC level. Other factors that may influence GC content include genome size and the extent of DNA methylation, which has been shown in vertebrate and hymenopteran insect like Polistes dominula (Mugal et al., 2015; Standage et al., 2016). The genomes of hymenopteran insect are in general characterized by relatively low GC levels and the gene conversion process and high recombination rates are hypothesized to contribute to GC bias (Branstetter et al., 2018). The smaller genome size and limited host species have been hypothesized to underline the extreme low GC content of the previously characterized aphid parasitoids (Dennis et al., 2020).
| Platform | Library size | Raw data (Gb) | Clean data (Gb) | Coverage (X) |
|---|---|---|---|---|
| Illumina | 350 bp | 25.2 | 18.6 | 118.5 |
| Nanopore | >10 kb | 11.8 | 10.9 | 69.5 |
| Hi-C | 400 bp | 82.7 | 80.0 | 509.9 |
| Methods | Statistic | Value |
|---|---|---|
| Nanopore assembly | Genome size (Mb) | 156.9 |
| Number of contigs | 136 | |
| Maximum contig size (bp) | 10,405,750 | |
| Contig N50 (bp)/L50 | 3,929,594/13 | |
| Contig N90 (bp)/L90 | 694,987/45 | |
| BUSCO (%) | 98.97 | |
| Hi-C assembly | Number of pseudo-chromosomes/scaffolds | 6/118 |
| Maximum scaffold size (bp) | 32,149,644 | |
| Scaffold N50 (bp)/L50 | 27,481,583/3 | |
| Total size (bp) | 155,454,520 | |
| BUSCO (%) | 98.90 |
| Features | Aphidius gifuensis | Aphidius ervi | Lysiphlebus fabarum | Cotesia vestalis | Diachasma alloeum | Fopius arisanus | Macrocentrus cingulum | Microplitis demolitor |
|---|---|---|---|---|---|---|---|---|
| Assembly level | Chromosome | Scaffold | Contig | Scaffold | Scaffold | Scaffold | Scaffold | Scaffold |
| Total length (Mb) | 156.9 | 138.8 | 140.7 | 178.5 | 384.4 | 153.6 | 132.3 | 241.2 |
| Number of scaffolds | 6 | 5743 | NA | 1437 | 3313 | 1042 | 5696 | 1794 |
| Scaffold N50 (kb) | 27,480 | 581.4 | NA | 2609.6 | 657.0 | 980.0 | 192.4 | 1,140 |
| Number of contigs | 13 | 12,948 | 1698 | 6820 | 24,824 | 8510 | 13,289 | 27,508 |
| Contig N50 (kb) | 3929 | 25.2 | 216.1 | 51.3 | 45.5 | 51.9 | 64.9 | 14.12 |
| Completeness (%) | 98.97 | 93.7 | 95.9 | 96.7 | 99.0 | 97.0 | 99.4 | 97.0 |
| GC content (%) | 26.5 | 25.8 | 23.8 | 29.9 | 38.3 | 39.4 | 35.6 | 33.1 |
| Repeat content (%) | 28.9 | 29.3 | 49.1 | 24.0 | 49.0 | NA | 24.9 | 36.2 |
| Number of protein-coding genes | 10,443 | 20,226 | 15,170 | 11,278 | 19,064 | 18,906 | 11,993 | 18,586 |
We generated 80 Gb of Hi-C filtered data (543,717,626 paired-end reads) to construct a chromosome-level assembly (Figure S2). After mapping these reads onto the draft genome, 138,972,005 unique PE reads were retained, including 113,641,396 valid interaction PE reads. Of the 136 contigs, 99.8% of the sequence length could be anchored onto six linkage groups, with the length ranging from 18.38 to 32.14 Mb, resulting in a scaffold N50 of 27.48 Mb (Table S2).
We assessed the completeness of genome at chromosome level using busco. We identified 1641 (98.97%) complete (single-copy genes: 94.75%, duplicated genes: 4.22%), 6 (0.36%) partial genes and 11 (0.66%) missing genes in the 1658 highly conserved Insecta data set (insecta_odb9) (Table S3). These results supported the high level of accuracy and completeness in the genome assembly.
3.2 Gene annotation and phylogenetic analysis
A total of 45.34 Mb of repeat sequences was identified, constituting 28.89% of the A. gifuensis genome (Table S4; Fig S3). DNA transposons and retroelements accounted for 4.85% and 3.37% of the genome, respectively. For retroelements, 2.15% were classified as long terminal repeats (LTRs), 1.07% as long interspersed elements (LINEs), and 0.14% as short interspersed elements (SINEs). Genome size usually appears to be positively associated with abundance of its repeat content (Bosco et al., 2007; Hartl, 2000; Maumus et al., 2015; Yin et al., 2018). Among all published genomes of parasitoid wasps, the genome of Macrocentrus cingulum, which is the smallest (132.4 Mb), contains 24.9% repeat elements (Yin et al., 2018), whereas larger genomes, such as those of Nasonia vitripennis (295.8 Mb), Diadromus collaris (399.1 Mb) and Diachasma alloeum (384.3 Mb), contain 42.1%, 37.0% and 49.0% repeat elements, respectively (Tvedte et al., 2019; Werren et al., 2010). However, an exception to this association can be found in Lysiphlebus fabarum, which has a much smaller genome size (140.7 Mb) but contains a high proportion of repeat sequences (49.1%) (Dennis et al., 2020).
Using three methods of gene prediction, 11,535 genes were annotated in the A. gifuensis genome (Table 4). The transcriptome-based method predicted fewer genes (6660 genes) compared to other wasps; for example, there are 11,278 genes in Cotesia vestalis, 15,328 genes in Diadromus collaris (Shi, Ma, et al., 2019; Shi, Wang, et al., 2019) and 19,597 genes in Microplitis demolitor (Burke et al., 2018), which were all predicted from RNA-seq data sets. Fewer putative genes with RNA-seq prediction may be affected by the limited life stage tissues because only male adults were used for the transcriptome sequencing in our study. The average gene length was 5921.89 bp, and the average CDs length was 1694.13 bp. The average exon number per gene, average exon length and average intron length were 5.36, 315.97 bp and 926.29 bp, respectively. In addition, approximately 90.53% of the genes (10,443 genes) could be functionally annotated (Table 5). We identified 98.90% of the BUSCO Insecta database (insecta_odb9) genes (single-copy genes: 96.30%, duplicated genes: 2.60%; Table S3) at protein level, further underlining the accuracy and completeness of gene predictions. Different types of noncoding RNAs were also annotated, yielding 846 tRNA, 130 miRNA, 100 rRNA and 77 snRNA (Table S6). The annotated gene set of A. gifuensis was compared with other parasitoid wasps (Table S5). The number of annotated genes was relatively low despite the high assembly quality and completeness of the A. gifuensis genome. In general, the existence of gap with shorter assembly scaffold or contig could increase the pseudogenes or false-positive annotations (Li et al., 2019).
| Prediction strategies | Software used | Total number of genes | Average gene length (bp) | Average CDS length (bp) | Average exons number per gene | Average exon length (bp) | Average intron length (bp) |
|---|---|---|---|---|---|---|---|
| De novo | AUGUSTUS | 14,256 | 4875.16 | 1562.98 | 4.74 | 329.67 | 885.35 |
| Homology | GeMoMa | 16,312 | 10,744.83 | 1638.44 | 4.90 | 334.25 | 2333.82 |
| RNA-seq | TransDecoder | 6660 | 4614.63 | 1613.78 | 4.64 | 347.58 | 840.84 |
| Final set | EVM | 11,535 | 5921.89 | 1694.13 | 5.36 | 315.97 | 969.29 |
| Database | Number | Per cent (%) |
|---|---|---|
| KOG | 7763 | 67.30 |
| KEGG | 6185 | 53.62 |
| NR | 10,332 | 89.57 |
| Swiss-Prot | 8877 | 76.96 |
| GO | 5746 | 49.81 |
| Total | 10,443 | 90.53 |
3.3 Orthology, synteny and phylogenetic relationship
Along with 13 other hymenopteran species, we identified 13,139 gene family groups in total and assigned 10,302 genes to 8261 gene families in the A. gifuensis genome using OrthoMCL. 3504 single-copy genes identified were employed to reveal the phylogenetic relationships among these species (Figure 2a). As shown in the phylogenetic tree, A. gifuensis was more closely clustered with Fopius arisanus than with Macrocentrus cingulum and Microplitis demolitor. These four species formed a clade of Ichneumonidae, which diverged from Chalcidoidea at approximately 194.6 Mya. The inter-Parasitica phylogenetic relationships were consistent with previous studies (Peters et al., 2017; Tang et al., 2019).
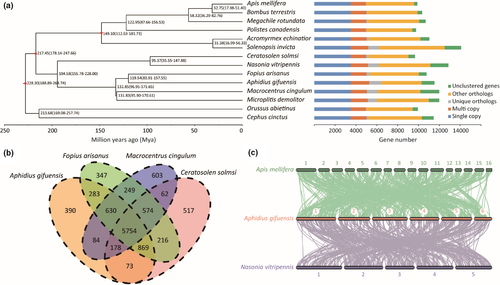
We compared the orthologous analysis between A. gifuensis and other three parasitoid wasps, Ceratosolen solmsi, Fopius arisanus and Macrocentrus cingulum. 5754 homology gene families were shared by the four species. A. gifuensis shared 7536 gene families with Fopius arisanus, higher than 6874 with Ceratosolen solmsi and 6646 with Macrocentrus cingulum (Figure 2b), that showed more homology between A. gifuensis and Fopius arisanus.
Syntenic relationships between A. gifuensis, Apis mellifera and Nasonia vitripennis showed a high level of collinearity among the three chromosome-level genomes from the Hymenoptera (Figure 2c). We defined a syntenic block as including at least three orthologous genes. In total, 542 syntenic blocks were found between A. gifuensis and Apis mellifera, and the gene number in these blocks ranged 4–23 with a mean of 5.97. 428 blocks were found between A. gifuensis and Nasonia vitripennis, with the same gene number range of 4–23 and a mean of 5.60. Syntenic analysis usually identifies putatively homologous genome regions by anchoring neighbouring gene pairs, which may influenced by differences in gene density, tandem duplication, gene transpositions and chromosomal rearrangements (Tang et al., 2008; Wang et al., 2012). In our analysis, A. gifuensis showed slightly higher synteny with Apis mellifera than Nasonia vitripennis, despite the closer phylogenetic relationship of A. gifuensis and Nasonia vitripennis, an observation which may be related to the above factors or to differing annotation qualities among these species.
3.4 Gene family expansion and contraction
Using café, we estimated the gene family expansion and contraction in A. gifuensis genome, compared with 13 hymenopteran species (Table S1) in the phylogenetic analyses. Significant expansions and contractions of gene families are usually related to the adaptive evolution of the species (Wu, Zhang, et al., 2019; Zhang et al., 2020). In the genome of A. gifuensis, a total of 405 and 663 orthologous groups were significantly expanded and contracted, respectively (Viterbi p < .05), compared to the most recent common ancestor (MRCA) of A. gifuensis and Fopius arisanus (Figure 3a).
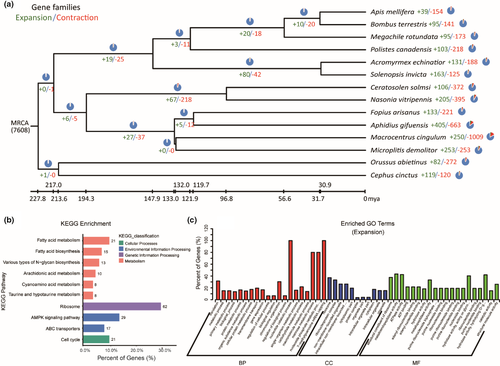
As shown by KEGG enrichment analysis among the expanded groups, several GO-terms were significantly over-represented: ABC transporters (02010, 17 genes, p = 7.76e-10), fatty acid biosynthesis (00061, 15 genes, p = 2.03e-7), AMPK signalling pathway (04152, 29 genes, p = 8.11e-7) and cell cycle (04111, 21 genes, p = .00141) (Figure 3b, Table S7). Based on GO analysis, some significantly enriched in metabolic process (GO:0008152, 316 genes, p = 1.11e-11), transmembrane transport (GO:0055085, 59 genes, p = 6.32e-6), chromosome organization (GO:0051276, 14 genes, p = .000112), signal transduction (GO:0007165, 18 genes, p = .00476), cellular macromolecule biosynthesis process (GO:0034645, 80 genes, p = .00679), ATP binding (GO:0005524, 114 genes, p = 1.22e-14) and transporter activity (GO:0005215, 63 genes, p = 3.13e-5) (Figure 3c, Table S8). Conversely, among the contracted gene groups, KEGG and GO analyses showed enrichments of olfactory receptor activity (GO:0004984, 5 genes, p = .00773), inorganic anion transport (GO:0015698, 3 genes, p = .00138), single-organism process (GO:0044765, 16 genes, p = .0006) and chromatin (GO:0000785, 2 genes, p = .00334) (Table S9, Table S10). In A. gifuensis, nutrients including proteins, carbohydrates and inorganic salts for larvae development are mainly absorbed from their hosts; therefore, cell transport and digestion are important.
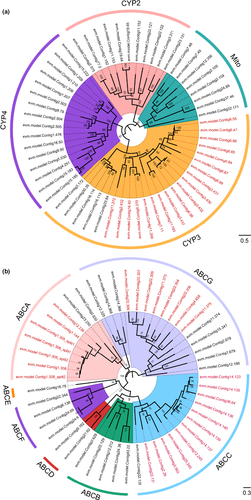
According to the results of café (Table S11), we manually annotated four gene families P450, a supergene family of enzymes found widely in eukaryotes, is involved in the metabolism of endogenous and exogenous compounds (pesticides, plant secondary metabolites, etc.) (Qiu et al., 2012; Scott & Wen, 2001). ABC transporter is a large class of transmembrane proteins, involved in the transport of xenobiotics (Merzendorfer, 2014; Wu et al., 2019). The two supergene families were well explored the insect adaptation to pesticide resistance and exogenous compounds (Cheng et al., 2017; Wan et al., 2019; Wu, Zhang, et al., 2019). In total, we identified 59 P450s and 50 ABC transporters (Figure 4a,b) and further analysed the number of genes in the subfamilies of P450 and ABCs in the genome of A. gifuensis. For P450s, the CYP3 and CYP4 subfamilies contained more genes than the CYP2 and Mito subfamilies. There were 19 CYP6 genes greatly expanded, based on CAFÉ results (Figure 4a). With increase biocontrol application and release of A. gifuensis, it has been proved that the sublethal toxicity of Imidacloprid had a significant negative effect on the lifespan, parasitic capacity of female adults and induced several genes expression, including central nervous system and detoxification system (CYP6a2, CYP49a3, POD and GST2) (Kang et al., 2018). For ABC genes, we identified seven subfamilies (ABCA-ABCG), and the subfamily ABCA, ABCC and ABCG expanded (Figure 4b). The expansion of CYP6 has been proven to be related to the resistance of pesticides and xenobiotic metabolism in several insects (Feyereisen, 2006; Müller et al., 2008; Wang et al., 2018). The subfamily ABCA, ABCC and ABCG was involved in cellular lipid transport, ion transport and toxin secretion (Guo et al., 2020; Wu, Zhang, et al., 2019). Therefore, the expansion of P450 and ABC subfamilies in the A. gifuensis genome could be related to resistance of pesticide and various metabolites of host.
Additionally, we manually identified venom protein genes of A. gifuensis, as these are one of the most important components for parasitoid wasps to ensure successful parasitism (Asgari & Rivers, 2011). We obtained 41 venom genes through a blast method querying the genome with sequences from several published wasps, such as Apis mellifera (Weinstock et al., 2006), Pteromalus puparum (Ye et al., 2020), Aphidius ervi and Lysiphlebus fabarum (Dennis et al., 2020). Among these genes, 9 venom carboxylesterase-6, 6 venom serine protease and 5 venom dipeptidyl peptidase showed expansion as identified in CAFÉ analysis (Table S11, Figure S4). The expanded venom serine protease may be closely related to the parasitic life history of this wasp, because the protease induces a lethal melanization response and exhibits fibrin(ogen)olytic activity in hosts (Choo et al., 2010). Although the venom proteins have been identified and reported in several parasitoid wasps (Colinet et al., 2013; Danneels et al., 2010; Dennis et al., 2020; Vincent et al., 2010; Ye et al., 2020), the different function of various venom genes still need further study.
Of particular note, we found a significant contraction in olfactory receptors (ORs) (Tables S9–S11), which have previously been hypothesized to be involved in locating hosts for parasitoid wasps (Wang et al., 2017). A previous study of A. gifuensis identified 66 ORs using transcriptome sequences (Fan et al., 2018). In this study, we found 80 putative ORs genes (Figure S5). The number was low compared to other hymenopteran parasitoids, with the exception of Macrocentrus cingulum, which has 79 OR genes (Ahmed et al., 2016). For example, there are 156 in Lysiphlebus fabarum, 228 in Aphidius ervi (Dennis et al., 2020) and 225 in Nasonia vitripennis (Robertson et al., 2010). The contraction of ORs may be associated with the limited host range of A. gifuensis.
4 CONCLUSIONS
In this study, we report a chromosome-level genome assembly of A. gifuensis, an important parasitoid biocontrol agent for multiple aphids. Genome assembly and annotation showed high completeness and continuity, with a contig N50 longer than most published genomes of parasitoid wasps. In addition, we identified genes putatively involved in insecticide resistance, parasitoid venom protein and chemosensing. The high-quality genome will provide a solid base for future studies on mechanism underlying parasitic biology, parasitoid–host interactions and currently population decline of this wasp in the artificial breeding, and will help to improve the commercial rearing and control efficiency of this wasp as an aphid control agent.
ACKNOWLEDGEMENTS
This work was supported by the China National Tobacco Corporation of Science and Technology Major Projects (Nos. 110202001036[LS-05] and 110201801023[LS-02]). We thank Prof. Xin Zhou, Dr. Shiqi Luo and Min Tang for their constructive comments on the genome assembly and annotation.
AUTHOR CONTRIBUTIONS
H. L., H. Y. and Y. Y designed the research. B. L., Z. F. prepared samples for genome and transcriptome sequencing. B. L., Z. D., S. W. and F. S. analysed data. B. L., Z. D., L, T. and H. L. wrote the manuscript. All authors reviewed the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
All raw data have been deposited into the NCBI Sequence Read Archive (SRA) database with a BioProject accession PRJNA615161 and a BioSample accession SAMN14446980. The whole-genome sequencing and transcriptome sequencing data are also available under Accession no. SRR11568632, SRR11568634, SRR11578829, SRR11578830 and SRR11577123–SRR11577219. In addition, the genome assembly and annotation information are available under the Accession nos of JACMRX000000000 and GCA_014905175.1. The version described in this paper is version JACMRX010000000.




