Development of Surface Chemical Strategies for Synthesizing Redox-Responsive Diatomite Nanoparticles as a Green Platform for On-Demand Intracellular Release of an Antisense Peptide Nucleic Acid Anticancer Agent
Abstract
Redox-responsive silica drug delivery systems are synthesized by aeco-friendly diatomite source to achieve on-demand release of peptide nucleic acid (PNA) in tumor reducing microenvironment, aiming to inhibit the immune checkpoint programmed cell death 1 receptor/programmed cell death receptor ligand 1 (PD-1/PD-L1) in cancer cells. The nanoparticles (NPs) are coated with polyethylene glycol chains as gatekeepers to improve their physicochemical properties and control drug release through the cleavable disulfide bonds (S–S) in a reductive environment. This study describes different chemical conditions to achieve the highest NPs’ surface functionalization yield, exploring both multistep and one-pot chemical functionalization strategies. The best formulation is used for covalent PNA conjugation via the S–S bond reaching a loading degree of 306 ± 25 µg PNA mg−1DNPs. These systems are used for in vitro studies to evaluate the kinetic release, biocompatibility, cellular uptake, and activity on different cancer cells expressing high levels of PD-L1. The obtained results prove the safety of the NPs up to 200 µg mL−1 and their advantage for controlling and enhancing the PNA intracellular release as well as antitumor activity. Moreover, the downregulation of PD-L1 observed only with MDA-MB-231 cancer cells paves the way for targeted immunotherapy.
1 Introduction
Cancer is a leading cause of death worldwide, with growing incidence and mortality rates year by year.[1] Advanced systems for controlled intracellular drug delivery can significantly benefit cancer therapy.[2] Long-term or extensive use of chemotherapeutics leads to side toxicity due to the lack of specificity between healthy tissues and tumors.[3] The tumor microenvironment has unique physiological features, such as acidic pH, hypoxia, increased redox potential, and enzymatic upregulation, thereby providing an opportunity to develop stimuli-responsive systems to release drugs with precise intratumoral localization.[4] Among these, responsiveness to redox stimuli has emerged as an appealing trigger due to the four-fold increase in the reducing glutathione (GSH) concentration in cancer cells compared to normal cells (i.e., 0.5–5 × 10−3 m in the cytoplasm; 2 × 10−6 m in the blood or extracellular matrices).[3, 5] This significant difference in the concentration of GSH between the healthy and tumor cells can prevent a premature drug release with a consequently reduced side toxicity. Since disulfide bonds (S–S) can be easily reduced in presence of GSH, nanosystems exploiting S–S bond as redox potential can achieve on-demand drug release.[6]
Hence, we developed a redox-responsive nanosystem based on diatom algae-derived natural silica for the controlled intracellular delivery of an antisense peptide nucleic acid (PNA) for cancer immunotherapy (Scheme 1).
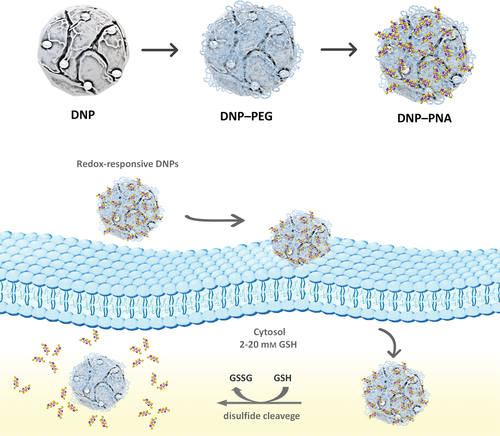
In recent years, there has been an increasing demand for safer, cost-effective, rapid, and greener methods for producing valuable nanostructures for biomedical applications to overcome the limitations arising from the production of synthetic materials.[7] Nature has developed elegant biologically based self-assembly synthetic routes to produce biomaterials with complex 3-dimensional (3D) porous structures and multifunctional properties. Among them, diatoms (unicellular photosynthetic algae) are a valuable eco-friendly source of nanostructured silica for biomedical applications due to the unique properties of their amorphous silica 3D-structured shell (so-called frustules), including large surface area (up to 200 m2 g–1), biocompatibility, chemical inertness, easy-to-tune surface chemistry, and stability.[7, 8] Moreover, it was proved that the unique architectures of diatoms cannot only provide the materials with a high loading degree capacity but also lead to efficient protection of the cargo until the point of release.The silica of diatoms is approved by the Food and Drug Administration (FDA), Generally Recognized as Safe for food and pharmaceutical production (GRAS, 21 CFR 182.90), and classified in the 3rd group of “Not classifiable as to its carcinogenicity to humans” by the International Agency for Research on Cancer (IARC).[9] In recent years, several in vitro and in vivo studies demonstrated the great potential of the frustules (whole or reduced in NPs) harvested from aquatic ecosystems or diatomaceous fossil sediments (diatomite) for drug delivery applications.[10] In particular, the effectiveness of diatomite NPs (DNPs) for gene therapy applications has been successfully demonstrated. The DNPs are able to deliver different small molecules by endocytic uptake and their accumulation mainly in the cytoplasmic region of cancer cells makes these systems suitable for antisense PNA transport to inhibit the intracellular transcription for cancer treatment.[11-15] PNA is a neutral charged artificial oligonucleotide mimetic molecule remarkably similar to DNA and RNA in intramolecular spacing and geometry.[16] PNA shows unique and improved properties compared to conventional oligonucleotides, including resistance to enzymatic digestion, higher stability combined with excellent hybridization affinity toward DNA and RNA, making it an attractive molecule for a wide range of biomedical applications.[17-19] Although PNAs act as effective antisense or antigene agents for inhibition of transcription or translation of target genes, their poor pharmacokinetics limits their therapeutic use. Therefore, there is a need to find valuable methods to enhance the access of PNAs into intracellular space, to increase the success of a PNA-based therapy.[20-23]
For this purpose, we developed an easy one-pot redox-responsive functionalization strategy to engineer natural DNPs for controlled antisense PNA intracellular delivery aiming to inhibit the immune checkpoint programmed cell death 1 receptor/programmed cell death receptor ligand 1 (PD-1/PD-L1) in cancer cells. PD1/PD-L1-mediated immune checkpoint is an important component of the immune escape mechanism used by tumor cells against the host immune system.[24] The activation of the PD1/PD-L1 signaling pathway is the mechanism by which tumors escape the recognition and killing by antigen-specific T cells, promoting tumor cell survival and metastasis. Clinical data demonstrated that the blockade of PD-1 signaling significantly enhanced antitumor immunity, producing durable clinical responses and prolonging patients’ survival.[25-27]
The DNPs were bioengineered with a polyethylene glycol (PEG) shell acting as gatekeeper for the controlled drug release, where the PEG and PNA were connected via a disulfide bond (S–S), cleavable upon exposure to a reducing environment.[28] Polymers are extensively used in the development of drug delivery systems to improve stability, biocompatibility, circulation half-time, and reduce adsorption of blood proteins, delaying the reticuloendothelial system's action (RES).[29-31] We explored different reaction conditions to obtain DNPs with the highest surface PEG functionalization yield and evaluated them by qualitative and quantitative techniques, such as one-dimensional (1D) solution-phase proton nuclear magnetic resonance (NMR), colorimetric assays, dynamic light scattering (DLS), transmission electron spectroscopy (TEM), and spectrophotometric analyses. The best NPs formulation was used for studies of PNA-conjugation, redox-triggered release, hemocompatibility, cells toxicity, cellular uptake, as well as activity on MD-MBA 231 triple negative breast cancer cells, A549 lung cancer, and U87 glioblastoma cells chosen for their high expression of PD-L1.
2 Results and Discussion
2.1 Multistep Chemical Approach for the Development of Redox-Responsive Nanosystems
A proper surface chemical functionalization of nanomaterials is paramount, especially for targeted drug delivery applications. The NPs’ surface is frequently functionalized to improve their physicochemical properties, enrich their functionalities, and affect their behavior in biological systems.[32-34] Therefore, the precise quantification of available surface functional groups on the NPs’ surface is fundamental to adequately control the surface chemical processes. To synthesize the redox-responsive DNPs with the highest surface functionalization yield, we explored different chemical conditions evaluating qualitatively and quantitatively the available functional groups by 1D solution-phase proton NMR, colorimetric assays, and DLS.
The first chemical approach was based on a conventional multistep procedure that introduces the desired reactive groups or functional moieties through independent and consecutive chemical reactions until obtaining the desired NPs. The bare NPs (DNPs–OH) were obtained by an ecofriendly method based on mechanical crushing, sonication, and acid purification of raw diatomaceous earth powder (see Materials and Methods, Figure S1, Supporting Information). The DNPs–OH were silanized by the chemical reaction between the triethoxy groups of aminopropyltriethoxysilane (APTES) and the −OH groups on the silica surface, thus introducing –NH2 groups needed for the covalent conjugation with the redox gatekeeper PEG molecules (5000 Da). The molecular weight (MW) of PEG molecules used for NPs PEGylation is fundamental to confer properties suitable for drug delivery applications. It was previously demonstrated that the silica NPs with PEG 5000 Da coating have higher aqueous solution stability than PEG 2000–500 Da coating; PEG with high MW has longer side chains than PEG with low MW, so it functions more effectively as a steric stabilizer and prevents adsorption of plasmatic proteins.[35]
The 1H NMR spectrum of APTES-modified DNPs solution in Figure 1A shows the characteristic peaks of the protons of the aminopropyl chain from 0.60 to 2.90 ppm (peaks a–c) and the absence of protons of ethoxy groups at 3.80 and 1.18 ppm (peaks d and e, Figure S2, Supporting Information), confirming the complete grafting of the APTES to NPs’ surface.[36] Proton NMR spectrum and chemical shifts of the free APTES as reference are shown in Figure S2 and Table S1 (Supporting Information).
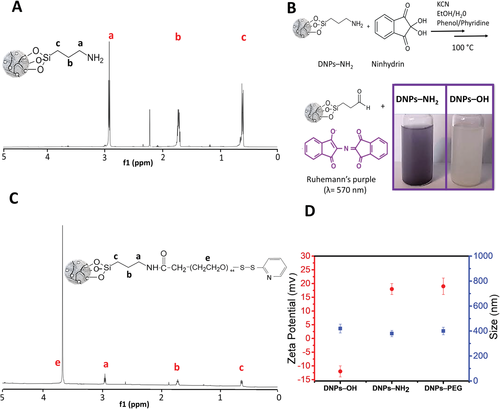
Then, we determined the number of APTES molecules onto NPs’ surface by quantitative NMR (qNMR). This technique is widely used for the quantification of small molecules in solution, and it has recently been explored in nanomaterials characterization to quantify chemical moieties on modified NPs, proving to be sensible, reproducible, and robust.[37-39] We quantified the DNPs silanization with the integration of the peaks area of 1H spectrum of the NPs solution by the internal standard fumaric acid (Figure S3, Supporting Information) and calculated 5.2 ± 0.5 µmol mg−1 of grafted APTES according to Equations (1) and (2) (see Experimental Section).
Subsequently, we used the ninhydrin assay for both qualitative and quantitative determination of the free –NH2 (see Materials and Methods, Supporting Information). The reaction of primary amines on DNPs with ninhydrin generates a colored product known as Ruhemann's purple measurable spectrophotometrically (Amax = 570 nm), whose color intensity is directly proportional to the content of accessible amines, which are needed in the further functionalization step, whereas the reaction of the bare NPs (DNPs–OH) with ninhydrin does not lead to any colored product (Figure 1B). Free APTES as standard in the range from 0.06 to 5 × 10−3 m was used for calibration curve purpose. The amount of accessible –NH2 on DNPs’ surface estimated by the analysis of the NPs supernatant resulting from ninhydrin assay, was 0.20 ± 0.06 µmol mg−1.
We then carried out the PEGylation of the DNPs–NH2 by the (N-hydroxysuccinimide) NHS esters of the heterobifunctional PEG molecules with the amine groups of the NPs at different conditions (see Experimental Section). The NHS esters are reactive groups formed by carbodiimide-activation of carboxylate moieties of PEG reacting with primary amines in physiologic to slightly alkaline conditions (pH 7.2–9) to yield stable amide bonds.
The 1H NMR spectrum of PEGylated DNPs shows a very weak signal of the large backbone (OCH2CH2)44 from 3.6 to 3.7 ppm, suggesting a low functionalization yield for both conditions, as confirmed by the qNMR analysis (0.002–0.005 µmol mg−1, corresponding to 1–2.5%, Figure 1C). Proton NMR spectrum and chemical shifts of the free PEG as reference are reported in Figure S4 and Table S2, Supporting Information).[40]
Next, we evaluated the hydrodynamic diameter and zeta potential of the DNPs before and after the chemical functionalizations by DLS. We observed a decrease of the NPs’ size from 420 ± 70 to 380 ± 50 nm after the silanization due to an increase of the DNPs’ surface repulsion forces passing from −12 ± 2 to 18 ± 2 mV, which confirmed the silanization process. However, no significant change in size and surface charge was observed after PEGylation (DNPs-PEG 400 ± 70 nm; 19 ± 2 mV), confirming the unsuccessful NPs’ PEGylation (Figure 1D).
2.2 One-Pot Chemical Strategy for the Development of Redox-Responsive Nanosystems
The low functionalization yield obtained by the standard functionalization strategies is not attributable to the PEGylation conditions used but to the susceptibility of APTES to hydrolysis upon exposure to the aqueous solution, high temperature, and sonication, leading to loss of functional amine groups (10–60%).[36] The hydrolytic degradation of APTES is mainly attributed to the primary amine being able to coordinate to the silicon center and catalyze hydrolysis via the formation of a stable five-membered ring.[41]
We used the above-mentioned steps as necessary procedures to remove adsorbed excess reagents or solvents, tailor the NPs in an aqueous solution usually used for bioconjugation processes, as well as for in vitro/ vivo tests, thereby causing a loss of functional amine groups.
Therefore, we developed an easy direct wet chemistry based on a one-pot strategy, allowing to silanize and PEGylate the DNPs’ surface as one step to prevent the loss of functional amine groups. This strategy consists in first synthesizing a complex of APTES–PEG, then used to directly functionalize the NPs avoiding intermediate purification steps that could mainly impact on functionalization yield.
We characterized the complex formation over time (up to 24 h) by 1H NMR and compared the obtained spectrum with free APTES one, paying attention to the 2.6–0.0 ppm region where occurring mainly proton signals change (Figure S5, Supporting Information). We observed a change in signals of the aminopropyl chain protons of the aminosilane due to the formation of an amide bond between the –NH2 of APTES and the activated –COOH of PEG, confirmed by the disappearance of signal a at 2.6 ppm, and the shift of b from 1.47 to 1.58 ppm. It is important to note the presence of the ethoxy group protons at 1.2 (e), validating that this chemical strategy preserves the APTES from hydrolysis, allowing then the silanization of the NPs.
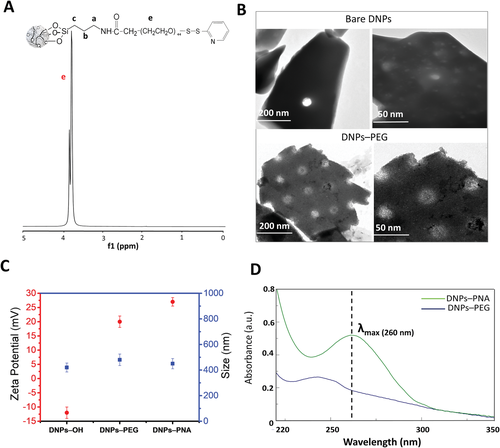
Subsequently, we functionalized the DNPs with the synthesized complex and evaluated the yield of reaction by qualitative and quantitative analyses. In Figure 2, 1H NMR spectrum of PEGylated DNPs showed an intense signal of protons of the large PEG backbone (OCH2CH2)44 from 3.7 to 3.9 ppm proving the successful NPs PEGylation, as confirmed by the qNMR analysis (6.5 ± 1 µmol mg−1, ≈80%).
We estimated the number of available thiol groups (–SH) on PEG-modified DNPs’ surface (DNPs–PEG) by colorimetric assay (ThermoFisher), after their treatment with dithiothreitol (DTT) reducing aqueous solution (20 × 10−3 m) to deprotect thiols from orthopyridyl disulfide (OPSS) groups (see Materials and Methods, Supporting Information).
In this assay, thiols reduce a disulfide-inhibited derivative of papain (papain–SSCH3), releasing the active enzyme (papain–SH). The enzyme cleaves the chromogenic papain substrate (L-BANPA) with the release of yellow nitroaniline chromophore (Amax = 405 nm) whose color intensity is directly proportional to the content of accessible thiols (Scheme S1, Supporting Information). We used L-cysteine at different concentration to create a standard curve and quantified 0.615 ± 0.065 µmol mg−1 of –SH available onto NPs’ surface for PNAs conjugation.
Subsequently, we evaluated the changes occurring in the NPs morphology after the one-pot modification by TEM imaging (Figure 2B). The analysis showed the typical irregular shape and porous morphology (10 nm < pores diameter < 50 nm) of the DNPs (bare DNPs), highlighting a dense polymeric layer onto NPs’ external surface and inside the pores due to the chemical modification (DNPs–PEG).[42] We further confirmed the success of the NPs’ functionalization by DLS analysis, resulting in an increase of the particles’ size from 420 ± 70 to 480 ± 85 nm and a change of surface charge from −12 ± 2 to 20 ± 5 mV (Figure 2C).
2.3 PNAs Synthesis and Conjugation to Redox-Responsive DNPs
Immunotherapy is radically changing conventional cancer therapy, and the immune checkpoint blockade strategy is the emerging target for developing innovative chemotherapeutics.[43] In particular, the targeting of PD-1/PD-L1 axis has shown impressive clinical benefit, even in hard-to-treat cancers.[44] To date, PD-1/PD-L1-directed antibodies are in clinical use. However, they are beneficial only for a small patients’ subset, expensive to produce and with risk of adverse effects thus limiting their use on a large scale.[45, 46] Therefore, novel therapeutics based on small molecules, easy to produce with unique physicochemical properties, such as PNAs, show potential with significant advantages compared to monoclonal antibodies. PNAs are nucleic acid mimic in which the phosphodiester backbone has been replaced with a polyamide backbone made up of repeating N-(2-amnioethyl)glycine units. The purine and pyrimidine bases are attached to the backbone and extend out in a conformation that is remarkably like standard oligonucleotides.[47, 48] To develop an effective antisense PNA to inhibit the PD-L1 expression, we used computational tools to select the specific mRNA region to target (NCBI Reference Sequence: NM_01 4143.4), as detailed in Experimental Section.
The synthesized PNAs (Table 1) were covalently bound onto the NPs’ surface by disulfide (S–S) bond formation due to exchange reaction among the sulfhydryl (–SH) side chains of the cysteine (Cys) residues of the PNAs and the OPSS of PEG-modified DNPs, respectively. Since having the thiols in a reduced form is mandatory for the success of the reaction, the PNAs were treated with an excess of reducing DTT prior the DNPs conjugation.[49] The NMR analysis of the DNPs–PNA showed very weak signals of amino protons of nucleobases (A, T, C) and aromatic amide protons of the PNA backbone (9.0–5.0 ppm); methyl protons of the A, T, C and the methylene protons of the PNA backbone (4-1 ppm), confirming the success of the bioconjugation (Figure S6, Supporting Information).[50] However, the backbone signal of the grafted PEG overbears the PNA signals, preventing a correct quantification the bonded-PNA (Figure S7, Supporting Information). After the PNA conjugation, the size of the DNPs did not change, contrarily to their surface charge which increased to 27 ± 3 mV due to the positive poly-lysine (poly-Lys) chain of the PNA (Figure 2C).
| Sample | Sequence |
|---|---|
| Wild-type PNA (PNA) | N-term-ACCATACTCTACCACATA-6Lys-Cys-C-term |
| Wild-type-FITC PNA (PNA*) | N-term-FITC-ACCATACTCTACCACATA-6Lys-Cys-C-term |
| Scrambled PNA (PNA-CTR): | N-term-ACATCATATCCAATCCAC-6Lys-Cys-C-term |
| Scrambled PNA-FITC (PNA-CTR*) | N-term-FITC-ACATCATATCCAATCCAC-6Lys-Cys-C-term |
Furthermore, we confirmed the successful PNA-conjugation onto the NPs surface by absorbance spectroscopy due to the appearance of an absorbance maximum at 260 nm, typical of the PNA, in the UV–visible (UV–vis) spectrum of DNPs (DNPs–PEG) after the conjugation with the PNA (DNPs–PNA) (Figure 2D). TEM analysis showed no changes in the nanosystems morphology after bioconjugation with the PNA; however, this result confirmed that the NPs exposure to the chemicals and procedures used in the biofunctionalization did not alter the chemical coating of the nanosystems (Figure S8, Supporting Information).
2.4 Drug Loading and Release Studies
As a result of its peculiar porous nature, diatomite NPs are ideal for the loading of a wide size range of molecules from small- to macromolecules. Moreover, it was previously demonstrated that the DNPs PEGylation did not impact negatively on the NPs porosity as well as loading capacity preserving a specific surface area (SSA) of 23.6 ± 0.1 m2 g–1. The reducing environment of tumors serves as an internal signal for the precise release of loaded drugs from redox-responsive nanocarriers.[51] Since the disulfide bond onto modified-NPs acts as redox-responsive linkage, we used this feature to release the bonded fluorescein isothiocyanate (FITC) labeled PNA molecules from the NPs surface and quantify it. The amount of PNA-loaded in the DNPs (i.e., the loading capacity) was determined by fluorescence analyses of the NPs supernatant after 48 h treatment with high DTT (20 × 10−3 m) concentration. The loading capacity of the DNPs was 0.0485 ± 0.004 µmol mg−1 (corresponding to ≈48% and 306 ± 25 µg PNA mg−1DNPs). Furthermore, when the particles were exposed to an excess of DTT (100 mm), no PNAs were further released.
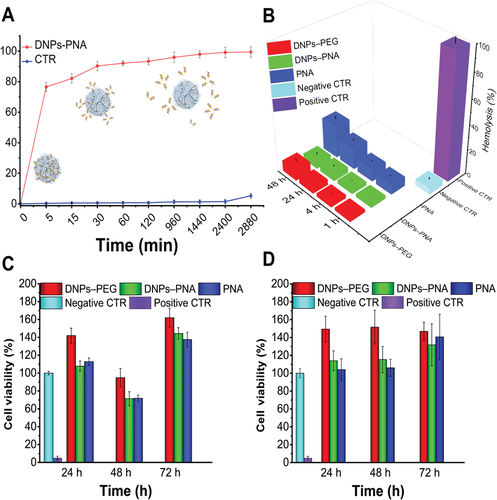
To investigate the release behavior of the redox-responsive nanosystems, we performed an in vitro release test under physiological conditions (phosphate buffered saline solution, PBS, pH 7.4), and in conditions mimicking the reducing tumor environment (PBS, DTT 20 × 10−3 m, pH 5.5). The tumor environment is highly reducing and hypoxic, with the intracellular GSH concentration at least four-fold higher than normal cells. This significant difference in the concentration of GSH between the healthy and tumor cells is relevant for the redox-triggered release into a specific site.[5] Figure 3A shows a complete release of PNA (≈98%, calculated with Equations (3) and (4)), with a plateau at 24 h, only for the DNPs treated with the reducing agent DTT (DNPs–PNA), contrarily, there was not much release (≈4%), in the absence of the DTT (CTR). Moreover, we also investigated the release behavior of the NPs in PBS at pH 5.5 mimicking the acid tumor environment and a PNA release comparable to the CTR was observed (data not shown). The obtained results confirmed that the disulfide bond cleavage controls the PNA release from the developed nanosystems and allows on-demand drug release at the tumor site.
2.5 Hemocompatibility Study
Blood is both the first contact for NPs administered intravenously and the gateway to reach target tissues or organs when administered via other routes. NPs without optimized and carefully crafted chemical surface properties can critically interact with the main cellular constituents of blood (i.e., red blood cell, RBCs) and influence both their structure and function with severe life-threatening conditions such as coagulopathy deep vein thrombosis (DVT) and disseminated intravascular coagulopathy (DIC). Therefore, hemocompatibility should be one of the foremost concerns in the design and development of NPs for therapeutic applications.[52]
The hemolytic activity of the DNPs–PNA was based on the quantification of percentage (%) of lysed RBCs after their exposure to the NPs at increasing incubation times (1, 4, 24, and 48 h) and concentrations (25, 50, 100, and 200 µg mL−1). We characterized the released hemoglobin spectrometrically at 577 nm and quantified the %-hemolysis with Equation 5 (see Materials and Methods, Supporting Information). The % of lysed RBCs after 48 h of incubation at the maximum concentration of modified-DNPs (200 µg mL−1) was 5 ± 2% for the DNPs–PNA, confirming the hemocompability of the developed redox-responsive nanosystems and their potential application for drug delivery applications. As control, the hemolytic activity of the free PNA molecules (0.01 µmol) is 30 ± 5% and 8 ± 2.5% for the DNP–PEG (200 µg mL−1), demonstrating that the bioconjugation onto NPs’ surface reduces the PNA hemotoxicity as well as improves the hemocompatibility of the PEGylated DNPs (Figure 3B).[9]
We ascribed the hemotoxicity of the free PNA to the thiol (–SH) of the Cys used to modify the molecule for the disulfide (S–S) bond formation with the DNPs–PEG. The hemotoxicity tested of the Cys alone (0.01 µmol) resulted much lower (2.5 ± 0.5%) than when bound to PNA because of the free carboxylic group (–COOH), which may lead to repulsion with negative charged RBCs’ membrane avoiding a possible thiol interaction with them (Figure S9, Supporting Information). In the bonded Cys–PNA, the –COOH is involved in peptide bond with Lys chain (used to give a positive charge to PNA for improving solubility) and lacking the repulsive force, the thiol freely interacts with the membrane of the RBCs and activates mechanisms that lead to the cells hemolysis.[53, 54] Finally, we also tested the hemotoxicity of a Lys chain-PNA sequence without Cys, resulting in blood compatibility (Figure S9, Supporting Information).
2.6 Cell Viability Studies
Another important characteristic in the development of drug delivery systems is the assessment of their safety when interacting with cells, which can help screening formulations, ensuring that only the safest one will be evaluated in vivo.[55] Although nanostructured silica from diatoms has been recently introduced in nanomedicine, to date, there are numerous in vitro and in vivo studies that demonstrated the biocompatibility and the safety of this emerging material.[10] The adenosine triphosphate (ATP) intracellular content, as an indicator of the physiological state of cells, was used for determining the viability of MD-MBA 231 triple-negative breast cancer cells and A549 lung cancer cells exposed to different DNPs and PNA concentrations (up to 200 µg mL−1 and 0.01 µmol, respectively) up to 72 h of incubation time (see Materials and Methods, Supporting Information). The physiological state of the cells incubated with the samples was compared with the negative control, represented by cells incubated with complete medium (10% FBS RPMI or DMEM, respectively). The modified-DNPs (DNPs–PNA) did not reduce cells viability on both cell lines, even at high concentrations and longer incubation times, confirming the safety of the DNPs for PNA delivery (Figure 3C,D).
As control, the cytotoxicity of the PEGylated DNPs (up to 200 µg mL−1) and free PNAs (up to 0.01 µmol) after 72 h was evaluated. The results confirmed the biocompatibility of the PEGylated NPs due to PEG coating, which positively affects the cells viability.[9, 56] In addition, PNA molecules resulted safe due to cell defence mechanisms, which overcome the potential toxicity of the free thiol group contrarily to the RBCs in which cell lysis is irreversible.[57]
2.7 Cellular Uptake Study
The cellular uptake was investigated by flow cytometry at two different incubation times (6 and 12 h) to evaluate the enhancement in the uptake given by the delivery of the PNA molecule with the diatomite-based hybrid nanosystems compared to the free PNA (see Materials and Methods, Supporting Information). A complete internalization was already observed after 6 h of incubation for the DNP–PNA particles, while free PNA further increased the internalization between 6 and 12 h (Figure 4A,B; Figure 5A,B).
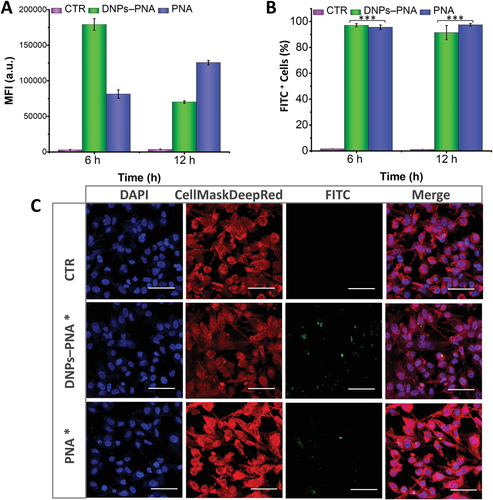
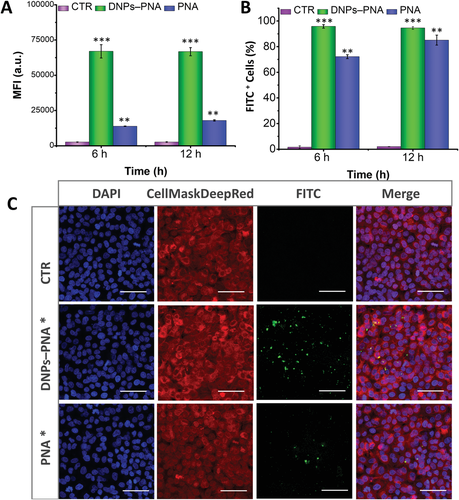
The cellular uptake evaluated by confocal microscopy imaging on both cancer cells up to 24 h of incubation with the labelled DNPs (DNPs–PNA*, 50 µg mL−1), confirming that these nanocarriers improved the internalization of the PNA molecules and increased their intercellular accumulation (Figures 4C,5C). In previous studies, the NPs modified with positively charged functions showed better cellular uptake than negative or neural charged ones due to electrostatic interaction with negatively charged cell membrane.[58, 59] Despite the PNAs being synthesized with the positive poly-Lys chain, the obtained results confirm the advantage of using NPs for the intracellular delivery of PNA, facilitating the cell membrane barrier crossing.
2.8 Activity Study
The PD-1/PD-L1 axis plays a key role in physiological immune homeostasis, and it is among the strategies through which cancer cells evade the immune system. The efficacy of the developed PNA-based nanosystems in downregulating PD-L1 was evaluated by flow cytometry analysis of the extracellular PD-L1 expression after cells exposure to DNPs–PNA 50 µg mL−1 up to 72 h.[60-62] For this study, MD-MBA 231 triple negative breast cancer cells, A549 lung cancer and U87 glioblastoma cells were chosen for their high expression of PD-L1 (Figure S10, Supporting Information).
Firstly, the downregulation of PD-L1 was functionally achieved only in MDA-MB-231 cells with a reduction of about 30% after 72 h (Figure 6A,B, and Figure S11A, Supporting Information). We initially hypothesized that a functional downregulation could be achieved only in cell lines expressing high basal level of PD-L1, ≈90% of MDA-MB-231 cells express PD-L1 on their membranes in the same conditions of the study (Figure 6B). The percentage of A549 cells expressing PD-L1 ranges between 40% and 60%, with a time-dependent increase in the expression in the assay conditions (Figure 6C,D, and Figure S11B, Supporting Information). Thereby, we chose the U87 cell line for its higher expression of PD-L1 (between 70% and 90%, with a time-dependent decrease in the assay conditions; Figure 6E,F, and Figure S11C, Supporting Information). However, the results obtained in U87 cells highlight that the basal expression of PD-L1 is not influencing the activity level of the PNA.
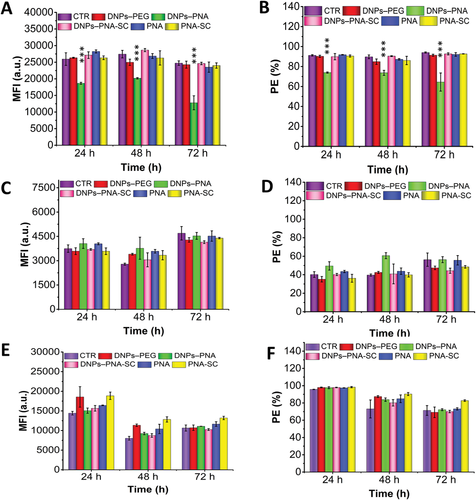
Importantly, the downregulation of PD-L1 on MDA-MB-231 was achieved only by the NPs conjugated to the wild-type sequence (DNP–PNA) proving that the NPs can effectively deliver PNA to the cells and the presence of the redox-responsive bond allows for the timely detachment of the PNA. Furthermore, the NPs conjugated to a scrambled PNA sequence (DNPs–PNA-CTR) built with the same components in random order does not show any effect, confirming the specificity of the PNA sequence produced towards PD-L1. Interestingly, in the case of A549 cells, the incubation with DNP–PNA particles increases the expression of PD-L1, while no statistically significant changes were detected in U87. However, further studies will be carried out to better understand these preliminary results. Despite the immune checkpoint blockade being an effective strategy for cancer treatment, the response to checkpoint blockade is not universal and varies significantly from tumor type. In the last years, compelling data have been provided on regulatory mechanisms of PD-L1, clarifying some mechanisms underlying primary and acquired resistance to PD-1/PD-L1 blockade conventional therapy. However, still many open questions need to be addressed in future research to support the emerging therapeutic approaches.[63-65]
3 Conclusions
Here, we describe the production of hybrid redox-responsive drug delivery systems using natural silica from diatomite that due to the unique physicochemical properties are ideal for controlled intracellular delivery of an antisense PNA. We properly designed and synthesized this promising therapeutic molecule to downregulate the PD-L1 expression on cancer cells and then inhibit the PD1/PD-L1 immune checkpoint for cancer immunotherapy. We developed a simple one-pot chemical strategy, which resulted superior to the conventional multistep strategy, to obtain NPs with the highest PEG surface coating needed to confer them stability, biocompatibility, and functional moiety for covalent PNA conjugation via the redox responsive disulfide bond. The drug loading study confirmed a conjugation of the PNA to the NPs of 0.0485 ± 0.004 µmol mg−1, corresponding to 306 ± 25 µg PNA mg−1DNPs. In vitro release study showed a redox-triggered release of the PNA (98%) in mimicking reducing tumor environment, while not much PNA release (4%) was observed in the case of a healthy cells mimicking environment. The hemocompatibility and biocompatibility of the nanosystems were tested on RBCs and MD-MBA-231, A549 cancer cells resulting safe up to 200 µg mL−1. Cell uptake studies have confirmed the advantage of using NP for the intracellular release of PNA whose internalization is low as a free molecule. The preliminary activity studies on different cancer cell lines (MDA-MB-231; A549 and U87) showed the downregulation of PD-L1 only on MDA-MB-231 by the NPs conjugated to the wild-type sequence (DNP–PNA), while no activity was observed in the case of NPs conjugated to a scrambled PNA sequence (DNPs–PNA-CTR), confirming the specificity of the synthesized PNA sequence toward PD-L1. Overall, the developed redox-responsive natural-based silica nanocarriers, easy to produce by a green method, with suitable physicochemical properties for on-demand drug delivery are promising for targeted therapy overcoming the drawbacks of conventional therapy. Since diatom-based silica has recently been explored in nanomedicine, it has not yet received official regulatory approval as a bioactive scaffold for drug delivery applications due to the lack of a consistent number of in vivo studies. However, this natural material is expected to contribute strongly to biomedical applications in a little while, considering its natural origin, unique physicochemical properties, biocompatibility, and low cost.
4 Experimental Section
Synthesis of Peptide Nucleic Acid Oligomers (PNA), Purification, and Analysis
To develop an effective antisense PNA, the 18-mer target sequence on the PDL-1 mRNA (NCBI Reference Sequence: NM_01 4143.4) was selected. First, the Uniprot database (https://www.uniprot.org) was consulted to identify the wild-type PD-L1 protein (Q9NZQ7) to select the corresponding mRNA tract (NM_01 4143.4) found in the NCBI dataset (https://www.ncbi.nlm.nih.gov). Within the whole transcript, the PD-L1 Open Reading Frame (ORF) was identified by the Reverse Translate Program, part of the Sequence Manipulation Suite tool (https://www.bioinformatics.org/sms2/rev_trans.html).
Into the ORF region, the most suitable sequence to target, also considering the appropriate characteristics of the PNA sequence to synthesize was identified. The following criteria had to be satisfied: (i) Isolate a high purine tract in the target mRNA to synthesize a complementary antisense PNA rich in pyrimidines and increase the yield % of the synthetic process; (ii) Design the hybridization process in the antiparallel orientation, considered the most stable and efficient form of interaction thermodynamically; (iii) Find nucleotide repeated motifs along the target and synthetic PNA strands to enhance the chance of a successful hybridization; (iv) Verify the lack of self-complementarity or self-dimerization, by using the OligoAnalyzer Tool (https://eu.idtdna.com) for both the selected mRNA tract and the complementary PNA sequences; (v) Demonstrate almost the absence of complementarity with the whole transcriptome in both parallel and antiparallel orientation, to avoid potential interactions with random RNA located in the cytoplasm by using the Basic Local Alignment Search (BLAST) Toolset in the BLASTn (nucleotide) interface (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The PNA oligomers with a 6-lysine chain (6Lys) to improve solubility and cellular internalization (Table 1) were synthesized by the 9-fluorenylmethoxycarbonyl (Fmoc) solid-phase strategy and purified by semipreparative HPLC analysis, according to the method described elsewhere.[66] The final pure products were characterized by electrospray mass spectrometry (ESI-MS) using a 4000 QTRAP mass spectrometer (ThermoFisher Scientific, Waltham, MA, USA) (Figures S12–S15, Supporting Information).
Prior to each conjugation step with the NPs, the sequences were also treated with an excess of DTT (PNA:DTT = 1:50 molar ratio) for 30 min under gently stirring to reduce eventually undesired disulfide bridges and purified by HPLC.[49]
One-Pot Synthesis of Redox-Responsive DNPs and PNAs Conjugation
The redox-responsive complex was synthesized by dissolving APTES and PEG (1:1 ratio, 7.5 µmol of each molecule) in 0.1 mL of DMF under constant stirring (800 rpm) for ON at RT. When elapsed the time, the bare DNPs (1 mg) were directly dispersed in the APTES–PEG/DMF solution under stirring, ON at RT. The NPs were centrifuged for 30 min at 15 000 rpm and the supernatant was discarded. The NPs (DNPs–PEG) were rinsed thrice with DMF, twice with MilliQ-water, and re-suspended in the same solvent. Finally, the DNPs–PEG (1 mg) with PNA wild-type (0.1 µmol, molar ratio NPs:PNA = 5:1) was conjugated in 0.5 mL of 40 × 10−3 m sodium phosfate, 2 × 10−3 m EDTA solution, pH 7.6 at 30 °C, ON under stirring. The DNPs–PNA were centrifuged for 30 min at 15 000 rpm and the supernatant was removed and the NPs were washed five times with MilliQ-water. The same bioconjugation procedure was used to functionalize the NPs with the different PNA sequences, as shown in Table 1.
NMR Spectroscopy Standard 1H NMR Experiments
The solution 1H proton NMR data was collected by a Bruker Advance III 700 spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany), operating at 700 MHz. The following samples, APTES (0.01–7.5 µmol) in 600 µL of DMF-d; OPSS-PEG-NHS (0.010–7.5 µmol) in 600 µL DMF-d; APETS/PEG complex (ratio 1:1, 7.5 µmol of each molecule) in 600 µL DMF-d; PNA 0.01 µmol in 600 µL DMF-d were all performed using standard 1H NMR protocol (128–256 scans, 2 dummy scans, 1 s relaxation delay). For qNMR, dried modified-NPs (0.1 up to 2 mg) were dispersed in 600 µL DMF-d or D2O, sonicated in an ultrasonic bath for 60 s, transferred to dry NMR tube (5 mm), and characterized by NMR (256 scans, 2 dummy scans, 6 s relaxation delay). The fumaric acid (10 mg in 1 mL of DMF) with resonance at 6.64 ppm was used as an internal calibrant and approximately 5–50 µL of the prepared solution the NPs sample was added before each analysis.
NMR spectra were processed by MestReNova software (Mestrelab Research, S.L., Spain). After Fourier transformation, the chemical shift was referenced by the residual signal of DMF-d7 used as solvent (a broad peak cantered at 8.03 ppm). The phase was corrected automatically first, followed manual adjustment for specific peaks if the automatic correction was not satisfying. The baseline was corrected by fifth-order polynomial fit with manually adjusted filter.
Drug Loading and Release
The drug loading capacity of the PNA-FITC modified-DNP (DNPs–PNA*) was determined by immersing the NPs (0.1 mg) into PBS buffer (1 mL, pH 7.4 and 5.5) without or with DTT (20 and 100 × 10−3 m) at 37 °C for 48 h under mild stirring. After the release test, an excess of DTT (100 mm) was added into the NPs suspension to completely release all PNAs. The NPs were removed by centrifugation, and the PNA* released in the supernatant was quantified by spectrofluorometer (FP-8250, Jasco Europe, Italy). All measurements were repeated at least three times. The amount of PNA was quantified using an external calibration method by comparing the fluorescence intensity of the supernatant at 520 nm with known concentration of PNA-FITC as standard.
Activity Studies
The activity of PNA in inhibiting the expression of PD-L1 was assessed by quantification of the expression of PD-L1 on the cell membrane by FCM. Briefly, 1 × 105 cells were seeded in 12-well plates and then placed back in the incubator for 24 h to allow for a complete attachment. The medium was then removed, and the samples were added at the concentration of 50 µg mL−1 in complete medium. The cells were then incubated with the samples for 24, 48, and 72 h. At each time-point, the samples were removed from the wells. Then, we washed the wells twice with PBS. The cells were detached from the wells with a scraper, transferred to a v-bottom 96-well plate and centrifuged at 300 g in an Eppendorf R5100 centrifuge for 5 min. The supernatant was discarded, and the cells were resuspended in 20 µL of TruStainX FcBlock solution (2 µL of TruStainX FcBlock plus 18 µL of PBS) and incubated at RT for 10 min. We then added 80 µL of Antibody solution to each of the samples, except for the blanks (1 µL PE anti human CD274 plus 79 µL of PBS). An amount of 80 µL of PBS was added in the blank wells. The cells were then incubated on ice at +4 °C for 20 min. The cells were washed twice with cold PBS, resuspended in 200 µL of cold PBS and analyzed with a BD Accuri flow cytometer equipped with C6 autosampler (BD Biosciences, USA). The data were then analyzed with floreada.io to quantify the mean fluorescence intensity and the percentage of PE positive cells, according to the gating strategy reported in Figure S15 (Supporting Information).
Statistical Analysis
Results of the assays are expressed as mean ± standard deviation (s.d.) of at least three independent experiments. Results were evaluated by means of one-way analysis of variance (ANOVA) with the level of significance set at the probabilities of *p < 0.05, **p < 0.01, and ***p < 0.001 using Origin 8.6 (Origin Lab Corp., USA).
Acknowledgements
M.T. acknowledges financial support from Programma Operativo Nazionale-Attraction and Internetional Mobility (PON-AIM) RTDA_L1 (Grant No. AIM 1873131-2). H.A. Santos acknowledges the Sigrid Jusélius Foundation, the Academy of Finland (Grant No. 331151) and UMCG Research Funds for financial support. The authors thank Dr. M. Pirozzi of the Euro-BioImaging facility, Institute of Protein Biochemistry (CNR), Unit of Naples for help with TEM analyses. The authors thank the Light Microscopy Unit, Institute of Biotechnology, University of Helsinki (supported by HiLIFE and Biocenter Finland) for confocal imaging. The authors thank Chiara Cassiano of the Mass Spectrometry Laboratory, Department of Pharmacy, University of Naples Federico II for mass spectrometry analysis.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




