NMR structure of hypothetical protein MG354 from Mycoplasma genitalium
Introduction.
Mycoplasma genitalium (Mg) and M. pneumoniae (Mp) are human pathogens with two of the smallest genomes sequenced to date (∼480 and 680 genes, respectively). The Berkeley Structural Genomics Center is determining representative structures for gene products in these organisms, helping to understand the set of protein folds needed to sustain this minimal organism. The protein coded by gene MG354 (gi3844938) from M. genitalium has a relatively unique sequence, related only to MPN530 from M. pneumoniae (68% identity, coverage 99%) and MGA_0870 from the avian pathogen M. gallisepticum (23% identity, coverage 94%), has no homolog with a determined structure, and no functional annotations.
Results.
The overall structure of MG354 (137 residues) was determined with a backbone root-mean-square deviation (RMSD) of 0.6 Å based on 850 structurally significant NOE restraints, 69 phi dihedral restraints, and 86 hydrogen bond restraints (Table I). A total of 40 N-HN residual dipolar couplings (rdc) were used to validate the model (see Methods). The structure consists of seven helices that fold into a single domain (Fig. 1). The most striking feature is the long central helix (H6), surrounded by the remaining six helices to produce a hydrophobic core. Within the CATH protein fold database,1 MG354 falls into the mainly alpha class and most closely resembles proteins with an orthogonal-bundle architecture.
| Restraints | |
| NOE upper distance limits | |
| Total | 850 |
| Intra-residue | 278 |
| Sequential (|i−j| = 1) | 238 |
| Medium range (1 < |i−j| < 5) | 213 |
| Long range (spanning 5 or more residues | 121 |
| Hydrogen bond restraints (two per H bond) | 86 |
| Phi torsion angle restraints | 69 |
| Distance restraint violationsa | |
| Residual DYANA target function | 1.4 ± 0.3 |
| Number of violations >0.2 (Å) | 3 ± 1 |
| Average maximum (Å) | 0.3 ± 0.1 |
| Torsion angle restraint violations | |
| Number >2.5° | 0 |
| Average maximum (degrees) | 0.02 ± 0.02 |
| van der Waals violations | |
| Number >0.3 Å | 1 ± 1 |
| Average maximum violation (Å) | 0.23 ± 0.08 |
| Coordinate precision (Å) | |
| Backbone atoms in helices (residues 5–15, 22–32, 46–54, 57–74, 86–90, 94–105, 119–135) | 0.6 ± 0.1 |
| Backbone (residues 5–135) | 1.1 ± 0.2 |
| Heavy atoms (residues 5–135) | 1.7 ± 0.2 |
| Procheck statisticsb (%) | |
| Residues in most favored regions | 64.5 |
| Residues in additional allowed regions | 28.7 |
| Residues in generously allowed regions | 5.8 |
| Residues in disallowed regions | 1.1 |
- a There were no distance restraint violations greater than 0.2 Å that occurred in eight or more structures. The maximum violation considering all structures and restraints was 0.6 Å.
- b Structures were not energy minimized.
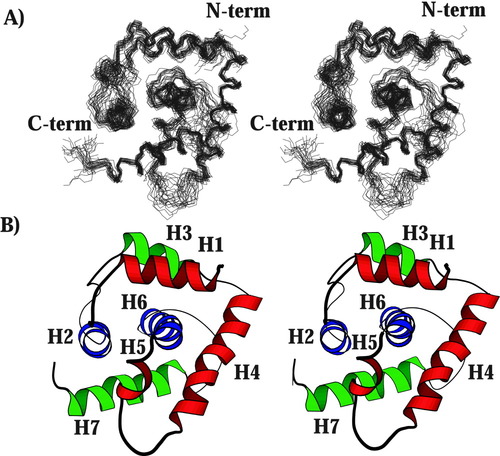
Structure of MG354. (A) Stereoview of the 26 final conformers. Residues 5–137 are shown. (B) Stereoview of a ribbon diagram of a representative structure generated with the program Molscript.23 Although not included in the calculations, the structures suggest that a disulfide bond forms between Cys-18 and Cys-99.
A search for similar folds was conducted with CE,2 Dali,3 and VAST,4 recently shown to be among the best at matching protein structures.5 Three energy-minimized coordinate sets from the 26-member family of NMR structures were used as templates. The three servers returned different sets of hits, each of which showed some similarity to MG354. For example, CE matched four contiguous helices (86 of 252 residues) in 1F5Q (chain B, γ-herpesvirus cyclin) to the last five helices in MG354 (H3 to H7) with an RMSD of 4.9 Å and a Z-score of 4.6. One helix in 1F5Q was paired with both H5 and H6. Combined, the match covered 62 and 8%, respectively, of the residues in domains 1 and 2 of the cyclin, as defined in the CATH database.1 Similarly, Dali matched four contiguous helices (75 of 508 residues) in 1GW5 (chain A, clathrin adaptor protein) to H1, H4, H6, and H7 of MG354 with an RMSD of 3.2 Å, and a Z-score of 3.9. However, the first helix of 1GW5 was oriented perpendicular to H1. Compared with these, VAST matched five helices (50 of 411 residues) in 1UX5 (formin homology-2 domain) to H1, H2, part of H3, H4, and H6 with an RMSD of 2.4 Å. In this case, the topology was somewhat different, because 1UX5 has an extra six-residue helix in between those that matched H2 and H3. Thus, all three methods were able to identify protein fragments that had some similarity to MG354, but none of the proteins contained all seven essential helices, and the matches were not strong enough to suggest similar functions.
In cases of low sequence and structural similarity, clues to function can sometimes be obtained by matching local structural patterns with those of known ligand binding sites and enzyme active sites. PROCAT6 classified MG354 as an acetylglucosamidase (E.C. 3.2.1.96) based on the locations of Glu-23 and Asp-26, and RIGOR7 classified MG354 as a lipase (E.C. 3.1.1.3) based on the positions of Glu-23, Asp-26, and Asp-41. The side-chains for all three residues are clustered together on the surface. Glu-23 and Asp-26 are at the edge of a 21 Å3 solvent-accessible cleft identified by the CASTp server.8 The cleft is defined by residues Gln-22, Leu-25, Asp-26, Leu-134, Asn-135, and Asn-137. However, PINTS,9 which uses a statistical score to filter out insignificant hits, returned no functional classification. The hydrolase and lipase predictions are intriguing, but the lack of consensus among the programs leaves the function of MG354 in doubt. Thus, because of its limited sequence and structural homology, the structure of MG354 represents a challenge for the further development of programs to predict function based on local structure comparisons.
Methods.
Protein Expression.
The gene for MG354 was cloned into the expression plasmid pSKB3 (a gift from Dr. Stephen Burley) and transformed into Escherichia coli strain BL21(DE3)pSJS1244 as described.10 Cells were harvested by centrifugation and lysed by sonication in 50 mM Tris-HCl (pH 8.0) and 300 mM NaCl. The supernatant was applied to a Ni-NTA column (Qiagen; Valencia, CA.), washed with the same buffer containing 15 mM imidazole, and eluted with a gradient up to 1 M imidazole. The His tag was cleaved with the tobacco etch virus (TEV) protease, and the protein was reapplied to the Ni-NTA column. The flow through was dialyzed against 20 mM potassium phosphate (pH 6.5), 100 mM NaCl, 1 mM EDTA, and 1 mM DTT and concentrated for NMR experiments. Final protein concentrations ranged from 0.75 to 1.2 mM.
NMR Spectroscopy.
NMR spectra were recorded at 25°C on a Bruker DRX 500-MHz spectrometer equipped with a triple-resonance cryoprobe and a Bruker DRX 600-MHz spectrometer equipped with a traditional triple-resonance probe. One three dimensional (3D) 13C-edited NOESY-HSQC spectrum was recorded at 800 MHz on a Varian INOVA spectrometer. Backbone 1HN, 15N, and 13C resonances were assigned using HNCO, HN(CA)CO, HNCA, CBCA(CO)NH, HNCACB, and 15N-separated NOESY-HSQC spectra recorded as described.11-13 1Hα and side-chain 1H and 13C resonances were assigned using HCANH, HBHA(CO)NH, H(C)CONH, (H)C(CO)NH, and HCCH-TOCSY experiments as described.11, 12 For a review of the experiments used, see Cavanagh et al.14 NMR spectra were processed using NMRPipe15 and analyzed using NMRView.16 A total of 92% of the resonances were assigned, including all amide protons except Lys-115.
Structure Calculations.
NOEs identified in 3D 15N-edited and 13C-edited NOESY-HSQC spectra and 4D 15N/13C-edited and 13C/13C-edited HMQC-NOESY-HSQC spectra were classified as strong (1.8–2.9 Å), medium (1.8–3.3 Å), or weak (1.8–5.0 Å) as described.11, 12 Phi angle restraints were derived from 3JHNHα coupling constants obtained from an HNHA spectrum.17 Angles were constrained to −60 ± 30° for 3JHNHα of less than 5 Hz and −170 ± 50° for 3JHNHα of greater than 8 Hz. DYANA18 was used for structure calculations. Manually assigned NOEs and torsion restraints were used to initially determine the fold. Subsequently, in-house software was used to assign NOEs by matching peak chemical shifts and filtering with distances in representative structures. Once the backbone RMSD decreased below 1.5 Å, hydrogen bond restraints were added for those amide protons with slow exchange (protection factors greater than 80) and short HN-O distances. Structures were viewed and analyzed with MOLMOL.19
To validate the structural model, 40 N-HN rdc were obtained from analysis of IPAP experiments20 recorded on isotropic and partially aligned (Pf1 phage) samples. At least two rdc values were measured for each helix. Using CNS,21 the best 20 of 200 structures had no distance or dihedral restraint violations greater than 0.5 Å and 5°, respectively, and the RMSD for N-HN rdc decreased from 4.6 ± 0.4 Hz (Qrdc22 = 0.48 ± 0.04) to 0.6 ± 0.2 Hz (Qrdc = 0.06 ± 0.01) with the additional restraints. The maximal residual coupling was 21 Hz. The mean structures calculated with and without rdc superimposed with a coordinate RMSD of 0.4 Å, indicating good agreement between the NOE, dihedral, hydrogen bond, and rdc data.
Acknowledgements
The authors thank Dr. Corey Liu for recording a 3D 13C-edited NOESY-HSQC spectrum at 800 MHz at the Stanford Magnetic Resonance Facility and Andrew Marshall of the J. Puglisi laboratory (Stanford University) for providing Pf1 phage. The authors are also grateful to Barbara Gold for cloning, Marlene Hernandez and Bruno Martinez for expression studies, and John-Marc Chandonia for bioinformatics assistance. Coordinates and structure restraints have been deposited in the PDB under accession number 1TM9 and the chemical shift assignments have been deposited in the BMRB under accession number 6244.




