Urinary Proteomics and Precision Medicine for Chronic Kidney Disease: Current Status and Future Perspectives
See accompanying commentary by Alberto Ortiz, https://doi.org/10.1002/prca.201900004
Abstract
Precision medicine is since long an ongoing refinement of classical medicine, integrating improved and more detailed pathophysiological understanding with rapid technological advances. In the heterogenous area of chronic kidney disease there seems to be a high potential for the improvement in treatment and prognosis for several causes, with new technologies under development, that are yet to be introduced in clinical practice. As in other medical disciplines, investigation of abundant peptide patterns (proteomics) has gained recent interest. Especially relevant for kidney disease, urinary proteomics may provide both improved diagnosis and, as reviewed here, also holds promise for personalized treatment in the future. So far, capillary electrophoresis coupled to mass spectrometry (CE-MS) is the most widely applied technique, and in addition to several cross-sectional and cohort studies, there is even an ongoing randomized controlled trial that will soon report on the concept used as a method of personalizing treatment. In addition, there is hope that urinary proteomics can turn into a “liquid biopsy,” replacing the invasive diagnostic procedure. The next couple of years will provide more answers on the topic.
1 Background
The area of chronic kidney disease (CKD) is a field of many causes and practices, and many conditions leading to CKD are not fully understood or curable. Along with a better understanding of pathology, there is a need for improved risk detection, for the determination of prognosis and for improved and personalized treatment. For such a development to happen, there will also be a need to enhance and implement a new technology to develop a more profound understanding of diseases and individual response to therapy ranging from beneficial effects to development of side effects. This would in turn require the nephrological community to accept the complexities that comes with individualized therapy, as has been seen in the latest developments in oncology and hematology, where mutation- or receptor-guided therapies are now commonplace.
Urinary proteomics is one such promising technology that could help with the development of personalized medicine, going from bench and currently on its way to bedside. In this review, we present the currently available clinical results and ongoing studies using this technology as well as put forward perspectives for the future place for this technology in the development and implementation of personalized medicine.
The concept of personalized medicine is rapidly growing into several different definitions. In this review, we define the concept as a way to tailor treatment guided by the physiology of an individual, the pathology of a disease, or even the individual response to a certain treatment.
2 Urinary Proteomics
Searching PubMed for “urinary proteomics” and “chronic kidney disease” reveals that the area is dominated by the capillary electrophoresis coupled to mass spectrometry (CE-MS) analytical approach. Only a few other publications investigate urinary proteomics patterns that contain more than a handful of conventional biomarkers. One such small study is from University of Michigan1 investigating glycoproteomic profiling as a mean to discover proteins in a case-control design, with urine sampled from six persons with CKD and six age-matched healthy controls. The lab used liquid chromatography electrospray ionization (ESI/LC) MS/MS analysis and a total of 122 glycoproteins were identified: 35 proteins unique to healthy controls, 8 were unique to CKD subjects, and 23 proteins were differently expressed in CKD as compared to the healthy subjects. In general, the identified proteins of interest were associated with inflammatory pathways. The authors state that this targeted technique may potentially be more specific for disease-related proteins as compared to other forms of urinary proteomic analyses in CKD where the urine samples also contains large numbers of abundant plasma proteins due to glomerular leakage. With only six persons included in the study, investigations with larger cohorts are obviously needed to establish the potential from this method to uncover CKD pathology and aid in precision medicine.
Using LC-MS/MS analysis, Caterino and Zacchia et al.2 found 73 and 66 proteins of interest in female and male patients with Bardet–Biedl syndrome (BBS), respectively. The patients had preserved kidney function, but proteinuria and renal structural abnormalities. Compared to healthy controls, these proteins, of which many are related to renal fibrosis, may help to improve understanding of the pathophysiology of a rare genetic disease and the authors hope that this can aid future preventive measures. Another study using this method found a number of urinary peptides to be related to early renal function decline in persons with type 1 diabetes in the first Joslin Study of the Natural History of Microalbuminuria in Type 1 diabetes.3 Interestingly, some of the peptide fragments in the urine were also found as corresponding proteins with increased expression in kidney biopsies, suggesting that biomarkers of early disease can be investigated from urine analysis.
In an interesting recent contribution, Yang et al.4 found urinary proteins specific for proliferative diabetic retinopathy in a cohort of 210 patients with type 2 diabetes. Using 2D nanoliquid ESI/LC tandem mass spectrometry, urinary haptoglobin was found to predict development of kidney impairment (estimated glomerular filtration rate [eGFR < 60 mL/min/1.73 m2]). This protein was the strongest biomarker available from 186 identified proteins in the urine.
A Polish case-control study5 in type 2 diabetes using liquid chromatography–tandem mass spectrometry found a decrease in urinary excretion of proepidermal growth factor, mannan-binding lectin serine protease 2, and basement membrane-specific heparan sulfate proteoglycan core protein (perlecan) associated with the progression of diabetic kidney disease. In addition, the authors found decreased urinary excretion of pancreatic amylase and deoxyribonuclease I indicating the presence of exocrine pancreatic insufficiency, a somewhat different finding as compared to most other cohorts.
Several methods for non-invasive biomarker discovery and investigation are evidently used in the literature. Only one comparative analysis was found in our literature search. Molin et al.6 compared capillary electrophoresis (CE) coupled to mass spectrometry (CE-MS) with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and concluded that CE-MS was superior in performance in terms of peptide resolution, with MALDI-MS having a reduced accuracy.
3 CE-MS
For the remaining part of this review, we will focus on urinary proteomics using CE-MS as this technique has developed farthest and broadest over the last decade, also in areas outside CKD.
The CE-MS is a powerful proteomic technology that enables the analysis of naturally occurring peptides and has been successfully applied in multiple clinical proteomics studies, as recently reviewed.7 The principle of the analysis lies in the coupling of a separation platform (CE) to a time of flight (TOF) mass spectrometer. During the first step of the analysis in the CE platform, the separation through fused silica capillaries is based on the differences in the electrophoretic mobility of the peptides, which is dependent on the charge and size of the molecule. Directly after the separation, the peptides are ionized and entered to migrate in the TOF mass spectrometer (MS), where the ion peaks are eventually detected. This technology showed very high reproducibility and accuracy,8 particularly in urine. In 2013, all necessary steps to carry out the urinary proteome analysis using CE-MS technology in kidney disease of humans were described.9 Rather than exploring only proteins in the urine, CE-MS can detect a wide range of peptides of many sizes and can provide of fragments from proteins present in the kidney or blood. The abundance of peptides places high demands on the bioinformatics used to handle and interpret the data. Support vector machine (SVM) is the analytical approach used to sort and present data to the researchers.
4 Molecular Studies
In one of the defining early studies of CE-MS in urinary proteomics,10 more than 20 clinical centers in Europe, Australia, and Asia collected urine samples from 3600 individuals, producing 5010 different peptides with distinct molecular signatures. From the urine peptidome database created, 273 biomarkers were combined in a pattern that was found to be associated with CKD. This pattern is now termed the CKD273 classifier and has been investigated in many different cohorts in an effort to expand and validate its applicability across cohorts and different stages of CKD.
5 Cohort Studies and Prediction of Disease
Whereas many studies are cross-sectional including different stages of renal disease,11 Zurbig et al.12 had the opportunity to investigate a mixed longitudinal cohort with 35 persons with type 1 or type 2 diabetes and normoalbuminuria at baseline with a long follow-up up to 20 years, and consecutive urine samples available. In a case-control design comparing subjects who stayed normoalbuminuric with subjects progressing to macroalbuminuria, it was demonstrated that the CKD273 classifier could predict the development of diabetic nephropathy (i.e., progression to macroalbuminuria) around 1.5 years before occurrence of microalbuminuria, by the early increase in numerous collagen fragments in the urine. This finding really set the stage for further investigations with the promise of a significant contribution to precision medicine. Diabetic nephropathy develops in approximately 40% of persons with diabetes and the possibility to predict its development opens the potential for early targeted intervention.
To further investigate these associations, several cohort studies were initiated. Roscioni et al.13 investigated the classifier and the potential to predict development and progression of microalbuminuria in type 2 diabetes in 44 cases and 44 controls with a follow-up of 3 years. Despite the small sample size, progression could be predicted on top of known predictors such as albuminuria and eGFR.
Pontillo et al.14 collected samples from 2672 persons with different CKD stages, of which 75% had diabetes looking to predict people with an eGFR decline of >5 mL/min/1.73 m2/year (fast progressors). In early stages of disease (eGFR > 70 mL/min/1.73 m2), the CKD273 classifier outperformed albuminuria in the detection of fast progressors, whereas albuminuria performed better in CKD patients with late-stage disease (eGFR groups 40–49 and <29 mL/min/1.73 m2). The performance of the CKD273 classifier was relatively constant in the eGFR groups 70–79, 60–69, 50–59, and 40–49 mL/min/1.73 m2. This study indicates that the use of this method may be most useful and have great advantage in early preventive precision treatment, selecting patients at increased risk. In later stages of diabetic kidney disease, it may however be more obvious to use albuminuria as marker of CKD. This also seems to be the case in the analysis of the Steno 2 study15 in which patients had micro- and macroalbuminuria, although this was a post hoc and potentially underpowered analysis. Recent developments may change this however, since there are now efforts to assign different subclassifiers of the CKD273 peptides to different eGFR strata. Rodriguez-Ortiz et al.16 showed that these specifically developed subclassifiers can outperform albuminuria and CKD273 at several stages of eGFR and may thus refine the clinical use of this approach. Especially relevant is the finding that rapid progressing eGFR decline can be predicted early, even in patients with preserved kidney function (eGFR > 60 mL/min/1.73 m2).
In 53 patients with mixed causes of CKD in a cohort with a mean follow-up of 3.6 years, Argiles et al.17 found that the CKD classifier was able to distinguish between patients (n = 15) that experienced an outcome of death or renal death and patients that did not. All patients with an outcome had a CKD273 score of more than 0.55.
Interestingly, Gu et al. could extend these findings to a general population, demonstrating association between CKD273 and continuous measures of renal function as well as CKD stages in 797 randomly recruited people.
Recently, in addition to the abovementioned studies predicting renal outcome, a detailed cohort of patients with type 2 diabetes and microalbuminuria examined and followed for cardiovascular disease, had the CKD273 classifier investigated and associated to calcium (CAC) score and independently to mortality.18
6 Comparison of the CE-ME with Standard Methods and Ability to Diagnose CKD
In a large cross-sectional cohort of 1990 patients with and without CKD, where 522 of the participants also had follow-up eGFR data available, Schanstra et al.19 demonstrated that the CKD273 classifier had a stronger association to baseline eGFR than albuminuria. During follow-up association to progression renal disease with decline in eGFR was also stronger with the classifier than albuminuria, lending the method both diagnostic and predictive quality across a mix of causes of CKD. Siwy et al.20 took this approach a step further when analyzing urine samples from 1180 subjects, and by SVM analysis, it was possible to discriminate between seven different etiologies of CKD. In the analyses the area under the receiver operating characteristic (ROC) curve ranged from 0.77 to 0.95, thereby quantifying that etiologic diagnosis can be improved by the use of this noninvasive approach. After the suggestion from Mischak21 that urinary proteomics can now be viewed as a “liquid biopsy,” the next logical step has recently been published by Magelhães et al.22 with an interesting paper comparing the analysis of 42 kidney biopsies with outcomes using urinary proteomics (CKD273 classifier). The authors found a significant association between urinary collagen fragments and the degree of fibrosis in the biopsy material. Interestingly, neither eGFR, urine albumin-to-creatinine ratio nor urine protein-to-creatinine ratio was associated to the degree of kidney fibrosis. The importance of these findings in relation to renal outcomes as progressive eGFR decline and development of end-stage renal disease remains to be established.
7 Trials Using Urinary Proteomics for Patient Selection and the Impact of Treatment on the Urinary Peptide Pattern
On the basis on the increasing amount of available cohort data supporting the use of urinary proteomics, the next obvious step is therefore to investigate prospectively whether the method can be used for selection of patients at risk of developing progressive diabetic nephropathy, and whether the selected patients at risk can then benefit from an early intervention. In addition, it is of interest to collect information on the effect of different treatments, that is, whether antihypertensive or antidiabetic treatments with documented renoprotective benefits affect the urinary peptide pattern and whether such changes reflect prognostic improvements.
In a post hoc analysis of the the Diabetic Retinopathy Candesartan Trials (DIRECT 2) study, in which patients with type 2 diabetes and normoalbuminuria were randomized to the angiotensin II receptor blocker candesartan or placebo with the intention of preventing retinopathy and nephropathy, urine samples were available for urinary proteomics analysis. Lindhardt et al. demonstrated that the CKD273 classifier, measured at baseline and thus before the intervention with candesartan, was an independent predictor of development of microalbuminuria, the renal secondary outcome of the study, improving risk prediction above usual clinical parameters with 14% relative integrated discrimination improvement (rIDI). In the original study, candesartan did not prevent the development of microalbuminuria in this cohort, and stratification on the basis of the CKD273 classifier did not improve the effect of the candesartan intervention, possible due to low numbers of patients in the high-risk group.
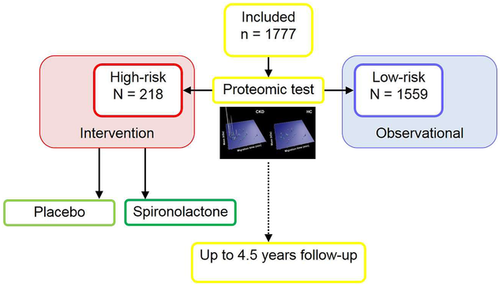
A major step in the long validation process of this method, and testing its usefulness in personalized medicine, is going to be the Proteomic prediction and renin angiotensin aldosterone system inhibition prevention of early diabetic nephropathy in type 2 diabetic participants with normoalbuminuria (PRIORITY) study.23 This a multicenter prospective study using the CKD273 classifier to select those at risk for the development of diabetic kidney disease among people with type 2 diabetes and normoalbuminuria. Patients will prospectively be identified with the “high risk” pattern (elevated CKD273 score) and compared for the rate of progression to those patients with the “low risk” pattern. In a second step, the “high risk” patients are randomized to spironolactone or placebo to test the hypothesis that it will be possible to prevent or delay the progression to microalbuminuria. The study population from 15 centers consists of 1777 patients of which 218 are “high risk” and have undergone randomization (Figure 1).24 The study is funded by the EU and is expected to report in 2019 and, being a prospective randomized study, will be a crucial step in the validation of urinary proteomics in this population both as a predictor of outcome and as well as a potential guide to intervention. The study poses two main questions: Is the CKD273 a useful test for detecting patients at risk and is a preventive intervention with spironolactone in the high-risk group effective? The number of participants singled out for randomized intervention is smaller than expected, and thus potential positive findings will need replication to prove the concept convincingly.
As discussed above, patients at risk for progression of CKD could be identified using urinary proteomic changes, but will improvement in the prognosis due to renoprotective intervention result in improvements or normalizations in the urinary proteomic pattern? This question was addressed in a cohort of type 2 diabetic subjects with microalbuminuria being treated with the angiotensin II receptor blocker irbesartan for 2 years in the Irbesartan in Patients with Type 2 Diabetes and Microalbuminuria (IRMA-2) study. This post hoc substudy of a part of the study cohort demonstrated that treatment with irbesartan was accompanied by a normalization in urinary proteomic pattern, and interestingly this normalization had a different time course than the reduction in urinary albumin excretion, underlining that the improvement in in the urinary proteome pattern was not just a reflection of the same processes being reflected by reduction in albuminuria which occurred faster and thus could reflect hemodynamic changes whereas the urinary proteomic pattern changes could be speculated to reflect more structural processes.25 A shorter study with another angiotensin II receptor blocker was also able to document improvements in the urinary proteomic pattern in diabetes patients.26Cherney et al.27 have made one of few published reports on the effect of modern antiglycemic treatment on the urinary proteomic pattern and the CKD273 classifier. In a post hoc analysis of an 8-week study with 40 persons with type 1 diabetes treated with the sodium glukose cotransporter 2 (SGLT2) inhibitor empagliflozin, it was found that the treatment significantly changed the amount of 107 peptides in the urine, indicating a more CKD protective peptide pattern in the urine after treatment. This is of special interest, since this class of treatment (SGLT2i) is attracting increasing interest for its potential renoprotective features with significant reductions in renal endpoints although so far only as secondary outcomes in studies primarily designed to be cardiovascular outcome studies,28 but a number of prospective dedicated renal studies are currently ongoing, of which the first (the Canagliflozin and Renal Endpoints in Diabetes with Established Nephropathy Clinical Evaluation, CREDENCE study) has been stopped because of positive outcome, although not yet presented.
It was also possible in patients with renal affection due to Fabry's disease to demonstrate changes in their urinary proteomic peptide pattern in the direction of normalization after treatment with enzyme replacement therapy30 in agreement with the concept that the treatment had impact on the pathogenetic pathway.
8 Prediction of Treatment Response
An important part of the potential of precision medicine is to predict treatment response by the use of pretreatment assessment of clinical, biochemical, or pathology testing. In many areas, this has already been implemented as for instance, the use of estrogen receptor marker testing in the treatment of breast cancer. Similar examples are now part of clinical practice in oncology and hematology but are not yet common in chronic disease areas as endocrinology and nephrology. In 111 patients with type 2 diabetes and hypertension Lindhardt et al.31 investigated whether the CKD273 could predict albuminuria-lowering response to spironolactone treatment in a post hoc analysis of a double-masked, randomized controlled trial. In the group of patients randomized to spironolactone a higher CKD273 score at baseline was associated to a larger treatment-induced reduction in albuminuria, which is considered a beneficial and renoprotective response. These results indicate that this method may have a place in precision medicine also for the prediction of response to treatment as a tool for individual tailoring of treatment.
9 Future Perspectives
The bulk of the data collected so far on the use of urinary proteomics is promising and indicative but the road to clinical implementation is long, both from a clinical and regulatory perspective.32 The utility of the method and critical appraisal of the accumulated evidence so far has been reviewed by Critselis and Lambers-Heerspink.33 They concluded that prospectively collected data is needed and that added value to available clinical information has to be demonstrated. It is clear the PRIORITY study23, 24 will provide the hitherto most comprehensive and prospective test of the concept, both for the selection of patients at risk and for the provision of early pharmaceutical intervention. Interpretation of the results will also help in the health economy considerations associated with widespread use and implementation as the testing is not without significant cost and will have to be balanced by significant future savings on diabetes-related morbidity.
One paper has looked into this question.34 The cost-effectiveness of the annual use of urinary proteomics for early assessment and modeled intervention in patients with type 2 diabetes, demonstrated in a Markov model that the use of the CKD273 classifier was more expensive than use of albuminuria measurement but did also yield more quality-adjusted life years (QALYs), when used in a high-risk population. This sort of analysis is crucial to model the overall benefit of early screening and can probably be performed again with higher perceived accuracy after the publication of the PRIORITY study.
Projects like the National Institute of Diabetes Digestive and Kidney Diseases (NIDDK) Kidney Precision Medicine Project in the United States and BEAT-DKD (www.beat-dkd.eu) in Europe/United States are aiming to link pathophysiological understanding of underlying and individualized disease mechanisms to prediction and intervention, and may be able to identify links between tissue data from biopsies including histology and transcriptomic analyses to biomarkers identified in urine or perhaps blood, using metabolomic or proteomic methods and potentially linking this to genetic data and clinical characteristics for a personalized profile allowing target intervention with minimal side effects and optimal outcome;35 however to what extent multiomics markers will add in clinical care to single omics markers or combinations of even single clinical markers still remains to be documented within CKD although the many initiatives are promising. A systems biology approach, combining multiomics information, has been suggested as the way forward to better combine and understand information provided by the molecular biomarkers at different stages of disease as well as providing the platform for design of individualized treatment. Mulder et al.36 provide an in-depth overview of an approach trying to integrate data from different candidate markers to be used for prediction of both risk and treatment response.
Taken together, after initial discovery stages, the area of urinary proteomics is currently undergoing strong and promising developments. Expecting continuing dedicated research efforts, the next stages will provide the necessary lessons needed before implementation in clinical practice can begin. This will depend on the results of the PRIORITY study and additional prospective clinical studies as well as a further detailed understanding of CKD pathology based on collected data. The day may soon come were kidney biopsies are abandoned and treatment is tailored based on urinary proteomic patterns.
Acknowledgements
The authors acknowledge support from the Novo Nordisk Foundation grant NNF14OC0013659 “PROTON Personalizing treatment of diabetic nephropathy” and the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 279277 (PRIORITY).
Conflict of Interest
F.P. reports having received research grants from AstraZeneca, Novo Nordisk, and Novartis; and lecture fees from Novartis, Eli Lilly, MSD, AstraZeneca, Sanofi, MSD, Novo Nordisk, and Boehringer Ingelheim; and having served as a consultant for Astra Zeneca, Bayer, Amgen, Novo Nordisk, and MSD. P.R. reports having given lectures for Astra Zeneca, Bayer, and Boehringer Ingelheim; and has served as a consultant for AbbVie, Astra Zeneca, Bayer, Eli Lilly, Boehringer Ingelheim, Astellas, Janssen, and Novo Nordisk, with all fees paid to Steno Diabetes Center Copenhagen, and has equity interest in Novo Nordisk.