Plasma Proteomic Signatures in Early Chronic Obstructive Pulmonary Disease
Abstract
Purpose
Chronic obstructive pulmonary disease (COPD) is characterized by progressive airflow limitation and abnormal inflammatory response of the lungs to inhaled noxious particles or gases. We used a proteomic approach with 2-DE followed by MALDI TOF-MS analyses in order to identify potential biomarkers in the early stages of the disease: global initiative for chronic obstructive pulmonary disease (GOLD) stage mild and moderate.
Experimental design
Blood plasma was collected from 43 patients with mild and moderate COPD as well as from 43 age- and sex-matched control subjects. Proteome analysis was based on 2D-Page followed by MALDI-TOF MS identifications. Validation was made on two significant proteins by western blotting.
Results
The analyses revealed 29 between-group differences in expressed spots, belonging to 20 unique proteins. These proteins are involved in inflammation (haptoglobin, Ig alpha-1 chain C), blood coagulation and complement pathways (prothrombin, complement 4-B, ApoH), oxidative stress (ceruloplasmin, vitamin D binding protein, and serotransferrin), and lipoprotein/lipid metabolism (apolipoprotein A-I, and apolipoprotein E).
Conclusion and clinical relevance
These results indicate that specific proteomic signatures can be detected and useful in terms of treatment selection and in early COPD patient monitoring.
1 Introduction
Chronic obstructive pulmonary disease (COPD) is a chronic disease characterized by a progressive airflow limitation that is not fully reversible, and an abnormal inflammatory response to toxic particles or gasses.1 COPD is currently the fourth leading cause of death in the world but is projected to be the third leading cause of death by 2020.2 COPD is characterized by obstruction of small airways, enlargement of air spaces and destruction of lung parenchyma, mucus production, and irreversible airway remodeling, resulting in respiratory airflow limitation and altered gas exchange.3 In industrialized countries, cigarette smoking accounts for most cases of COPD, while in developing countries other environmental pollutants play a significant role.4 This suggests a complex interaction between environmental and genetic factors. It is believed that the driving force in the pathogenesis of COPD is an abnormal inflammation in the smaller airway compartment with upper or lower production of inflammatory mediators in the bronchoalveolar lavage fluid (BALF), sputum, and lung tissues from patients with COPD.15 Bronchioles are obstructed by fibrosis and there is a concomitant infiltration of macrophages and T lymphocytes (CD8 + T cells) and an increased number of neutrophils.5-7 Macrophages and neutrophils release proteinases that break down connective tissue in the lung parenchyma, resulting in emphysema, and stimulate mucus secretion.8 Oxidative damage can also lead to the activation of antiproteases, such as a α1-antitrypsin and secretory leukoprotease inhibitor, amplifying inflammation and proteolytic injury.3 Although COPD may be suspected in the presence of persistent cough, sputum production, and exertional dyspnea, the diagnosis is based on spirometry measurement with a post bronchodilator forced expiratory volume in 1 s over forced vital capacity ratio (FEV1/FVC) below 0.7, according to the global initiative for obstructive lung disease (GOLD).9 COPD can be categorized into mild, moderate, severe to very severe according to FEV1 values, although correlation with clinical symptoms is weak in the early stages.10, 11 Spirometry reflects disease severity rather than disease activity, and its measurements strictly depend on patient's compliance, physician's expertise, and data interpretation.12 The limitations of pulmonary function tests have encouraged the search for circulating biomarkers of the disease. Two-dimensional gel electrophoresis (2-DE) is an easy and widely proteomic separation technique used to analyze differentially expressed proteins in biological samples. By proteomics analysis can be simultaneously identified multiple proteins associated with different disease states13; that has been applied to investigate the protein changes in the lung tissue,14, 15 BALF, and induced sputum of COPD patients.16-21 However, the identification of specific proteomic signatures from plasma might provide a simpler, and safer, approach. Nevertheless, the proteomic investigation of human plasma, especially by 2-DE has proved to be challenging, because abundant proteins, such as albumin and IgG, mask the detection of low-abundance proteins, some of which might potentially be relevant to particular disease states. In this paper, we performed a proteomic approach using 2-DE followed by MALDI TOF-MS analyses in order to highlight potential biomarkers in mild and moderate COPD that can be used as indicators of the disease progression. Since most studies have focused on the more severe stages of COPD, we investigated protein behavior in the first stages of the disease (GOLD stage mild and moderate) in order to identify biomarkers of early disease onset.
Clinical Relevance
Chronic obstructive pulmonary disease (COPD) is a complex disease characterized by a progressive airflow limitation not fully reversible and an abnormal inflammatory response to toxic particles or gasses. While most of the proteomics studies on plasma samples have focused on the more severe stages of the disease, where the protein changes are supposed to be more relevant, this work aimed at discovering proteomic signatures at the early stage of the disease (GOLD stage mild and moderate). The 2D-page analysis revealed important detectable signatures in proteins related to inflammation, regulation of blood coagulation, lipid metabolism, and oxidative stress. Among them haptoglobin, fibrinogen, ceruloplasmin, Ig alpha-1 chain C, complement C4-B, prothrombin, serotransferrin, Apo A-I, and vitamin D binding protein appear as being differentially expressed with a general increase of acute phase proteins and a concomitant depletion of proteins with antioxidant properties. Furthermore, some proteins show an increase only in the mild COPD group suggesting a temporary imbalance of proteins that in the first stages of the disease try to counteract the negative effects of the increasing inflammation status. The protein changes identified so far could help in the identification of novel biomarker panels that may be useful in terms of treatment selection and patient monitoring.
2 Experimental Section
2.1 Subjects and Design of the Experiment
We studied 43 COPD patients (mean age 74.8 ± 5.9 years, range 52–85 years), 29 with mild, 14 with moderate disease, and 43 age- and sex-matched healthy controls. The exclusion criteria were the presence of concomitant inflammatory disease such as autoimmune disorders and infections, cancer, liver, kidney, and heart disease. None of the patients involved in the study had any history of asthma or atopy. All subjects were recruited from the Respiratory Unit of the University of Sassari. Participants underwent physical examination, chest radiographs, routine blood tests, and respiratory function tests including FVC, FEV1, and FEV1/FVC ratio. All clinical data, including age, body mass index (BMI), and demographic information, for example, occupation and smoking status, were obtained through a structured questionnaire. COPD patients with significant symptom deterioration within the last 3 months, suggestive of disease exacerbation, were not enrolled. In the design of the experiment the same proportion of current, never, and ex-smokers has been used, in particular: Controls (CS = current smokers, 7%; NS = never smokers, 32.6%; ES = ex-smokers, 60.4%), mild COPD (CS = current smokers, 3.4%; NS = never smokers, 31.1%; ES = ex-smokers, 65.5%), and moderate COPD (CS = current smokers, 7.1%; NS = never smokers, 14.2%; ES = ex-smokers, 78.6%). No patient received inhaled corticosteroids (ICS) within 4 weeks prior to the study, or was on long- or short-acting β-agonists (LABA or SABA) or long-acting muscarinic antagonist (LAMA) at the time of the assessments. Diagnosis and severity of COPD was assessed according to smoking history, physical examination, respiratory symptoms, and spirometric results based on the GOLD criteria.22 Diagnosis of COPD required confirmation of airflow limitation that is not fully reversible (post-bronchodilator FEV1/FVC ratio < 0.7). Table 1 shows the classification of COPD severity based on spirometric values. This study was approved by the Institutional Local Ethics Committee (Azienda Sanitaria Locale n 1 di Sassari (Italy) (prot. 2175/CE, 21/04/2015), and was in accordance with the principles of Declaration of Helsinki. All subjects provided written informed consent.
| Characteristics | Controls (n = 43) | COPD mild (n = 29) | COPD moderate (n = 14) |
|---|---|---|---|
| Age (years) | 73.4 ± 6.9 | 75.4 ± 4.8 | 73.4 ± 7.7 |
| Sex (F/M) | 9/34 | 7/20 | 2/12 |
| BMI (kg m−2) | 26.4 ± 3.6 | 27.4 ± 3.4 | 27.4 ± 4.4 |
| Current smokers | 3 (7%) | 1 (3.4%) | 1 (7.1%) |
| Never smoked | 14 (32.6%) | 9 (31.1) | 2 (14.2%) |
| Ex-smokers | 26 (60.4%) | 19 (65.5%) | 11 (78.6%) |
| FEV1 (L) | 2.64 (2.33–3.09) | 2.30 (1.70–2.45)a) | 1.44 (1.36–1.83)a) |
| FEV1 (% predicted) | 111 (103–123) | 93 (87–98)a) | 63 (51–73)a) |
| FVC (L) | 3.26 (2.91–3.80) | 3.24 (2.67–3.51) | 2.41 (1.97–2.98)a) |
| FVC (% predicted) | 108 (97–120) | 99 (96–101)b) | 77 (60–87)a) |
| FEV1/FVC | 79.8 (78.4–83.5) | 69.1 (68.2–69.5)a) | 64.9 (60.2–69.0)a) |
- a p < 0.001 versus baseline.
- b p < 0.5.
2.2 Sample Processing
Whole blood was collected in EDTA-coated tubes. The red cell fraction was separated by centrifugation at 2500 × g at 4 °C for 15 min and the clear plasma supernatant stored in aliquots frozen at −80 °C. For the proteomic analyses, three plasma pools were prepared for the three subject groups (controls, mild, and moderate COPD patients). Plasma was depleted from the two most abundant proteins albumin and immunoglobulins with a commercial kit PROTBA (Sigma). The protein content was quantified using the Pierce660 Reagent (LifeSciences) which is based on a Lowry modified protein assay.
2.3 Two-Dimensional Gel Electrophoresis (2-DE) and Visualization
After protein extraction and quantification, the first dimension of the 2D-SDS PAGE was carried out with Ready Strip immobilized pH gradient (IPG) strips 17 cm with nonlinear pH 3–10 (BioRad). The strips were rehydrated with 300 μg of protein sample in rehydration solution (Urea 8 M, Thiourea 2 M, 4% CHAPS, DeStreak reagent, 10 mM DTT(1,4-Dithiothreitol), 1% carrier ampholyte 3–10, and 0.05% bromophenol blue) for 20 h. After rehydration, proteins were separated in the first dimension for a total of 72 000 Vh. After isoelectric focusing, the IPG strips were equilibrated in a two-step equilibration protocol. The equilibration buffer contained 50 mM Tris-HCl pH 8.8, 6 M urea, 20% v/v glycerol and 2% w/v SDS. For the first equilibration step, 2% DTT was added to the equilibration buffer and 2.5% iodoacetamide for the second one. After equilibration, proteins were separated in the second dimension by SDS-PAGE on 8–16% gradient acrylamide gels using the Protean Multicell (BioRad) apparatus.23 Gels were run for 15 min at 25 mA and then for ∼5 h at 50 mA in Tris-glycine-SDS running buffer.
2.4 Image and Statistical Analyses
The analytical gels were stained with Sypro Ruby fluorescent solution (Invitrogen). Gels were fixed in 50% methanol, 7% acetic acid in two steps of 30 min each, incubated overnight with Sypro Ruby gel stain and washed for 30 min with 10% methanol and 7% acetic acid. Preparative gels were stained overnight with Coomassie Brilliant Blue R-250 (CBB R-250) solution (0.05% w/v CBB R- 250, 50% v/v methanol, 10% v/v acetic acid). Gels were scanned using ChemiDoc XRS (BioRad Hercules) for Sypro Ruby stained gels or GS-800 densitometer (BioRad) for CBB R-250 staining. Gel images were analyzed with PDQuest 2D Analysis Software (BioRad) for spot matching and gel comparisons.24 In order to exclude artefacts and false positives we manually confirmed both matching and data quality of all the spots. The protein spot mean intensity values (OD, optical density) were normalized using the total density in gel image method available in the software. The quantity table with normalized data was exported to a spreadsheet file and submitted to statistical analyses using the software SPSS v.21. Only those spots with significant change (p-value ≤ 0.05 by Student t-test) were selected for MS analyses.
2.5 In-Gel Digestion of Proteins
Protein spots were manually excised from the preparative gels, washed three times for 15 min with 50 mM NH4HCO3 pH 8 and with acetonitrile (ACN) for another 15 min. The gel pieces were reduced with 10 mM DTT in 50 mM NH4HCO3 for 45 min at 56 °C and alkylated with 55 mM iodoacetamide in 50 mM NH4HCO3 for 30 min at room temperature in the dark. After incubation several washes were made with 50 mM NH4HCO3 pH 8 and ACN until the blue dye disappeared. After total evaporation of the ACN the gel pieces were hydrolyzed with 10 μL of trypsin (10 ng μL−1) in 50 mM NH4HCO3 pH 8. After 2 h of incubation on ice the supernatant was removed and the gel pieces resuspended in 50 mM NH4HCO3 pH 8; the samples were incubated overnight at 37 °C. After digestion, the protein peptides were acidified with 20% TFA, evaporated in a vacuum centrifuge and resuspended in 5 μL of 2% TFA.
2.6 Protein Identification by Mass Spectrometry
MS analysis was performed on a MALDI-TOF mass spectrometer micro MX(Waters). One microliter of tryptic peptide solution of each digested spot was mixed with an equal amount of the matrix α-cyano-4-hydroxycinnamic acid (CHCA), prepared in 0.2% TFA and in 70% ACN, applied on a MALDI plate and dried at room temperature. All spectra were acquired in reflectron mode, positive ion, and mass range from 800–4000 Da. Ionization was performed by irradiation with a nitrogen laser (wavelength, 337 nm) operating at 20 Hz. For matrix suppression, we used a high gating factor with signal suppression of up to 800 Da. The spectra were calibrated externally resulting in mass accuracy better than 100 ppm, using ProteoMass Peptide and Protein MALDI-MS Calibration Kit using the following mixture of peptides: Bradikinin fragment 1–7 (m/z 7 573 997 Da), Angiotensin II (m/z 10 465 423 Da), P14R synthetic peptide (m/z 15 338 582 Da), ACTH fragment18-39 (m/z 24 651 989 Da), Insulin oxidized β chain (m/z 34 946 513 Da). All spectra were analyzed using the Mass Lynx v 4.1 software (Waters). Protein identification was performed by peptide mass fingerprinting (PMF) using MASCOT software searching (http://www.matrixscience.com). Search parameters were restricted to Homo sapiens taxonomy using the Swiss-Prot database. Enzyme selected was trypsin, with up to one missed cleavage permitted. Carbamidomethylation of cysteines was selected as a fixed modification; Gln–>pyro-Glu (N-term Q), Oxidation (M) as variable modifications. Protein mass was unrestricted, and peptide mass tolerance typically set at ±150 ppm. Mass values were entered as monoisotopic MH+.
2.7 Western Blot Validation
For monodimensional western blot, 25 μg of depleted proteins from each pooled sample group was loaded on each lane in triplicate and resolved on SDS-page on 10% gel. Resolved gels were electroblotted on nitrocellulose membranes (Biorad) at 300 mA for 1 h. Membranes were incubated with blocking solution (PBS, 0.05% Tween 20, 5% dry milk) for 1 h at room temperature followed by overnight incubation with anti-vitamin D binding protein (VDBP) ab81307 rabbit monoclonal antibody (abcam) at a dilution of 1:10 000 or rabbit polyclonal anti-fibrinogen antibody ab83477 at a dilution of 1:3000. After 30 min washing steps with PBS-Tween 20, membranes were incubated for 1 h at room temperature with anti-rabbit (abcam) or anti-goat (Santa Cruz), HRP conjugated secondary antibody both at 1:10 000 dilution. After washing, membranes were revealed with enhanced chemiluminescence (Clarity western ECL Biorad). Membrane images were acquired with the ChemiDoc XRS (BioRad Hercules) and the staining by red ponceau was used as a protein-loading control. ImageLab software (BioRad) was used for densitometric analyses. The OD of the proteins was expressed as a percentage of the volume. Statistically significant differences were calculated by Student t-test (p ≤ 0.05) with the software SPSS v.21.
2.8 Protein–Protein Interaction Network Analysis
Differentially expressed proteins were mapped onto STRING v10 to build protein–protein interaction network with an interaction score higher than 0.4. STRING v10 is a database of known and predicted protein–protein interactions as derived from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other (primary) databases.25
3 Results
3.1 Characteristics of Patients Participating in the Study
Patient characteristics are summarized in Table 1. There were no significant differences in terms of age, gender, or BMI. The differences in the ratio FEV1/FVC was used to classify participants in the three groups according to the GOLD.9
3.2 2-DE Experiments and Mass Spectrometry Identifications
To examine the differential expression of proteins between mild or moderate COPD groups and the control group proteomic analysis was performed on plasma samples using high-resolution 2-DE. Average spot numbers were 408, 394, and 382 for control, mild, and moderate COPD, respectively. The spots showing significant differences (p < 0.05 Student t-test) in normalized intensities between mild or moderate COPD and the control group were 35, 37, and 23, respectively. Among them, 29 spots were identified with MALDI-TOF technology. Figure 1 shows a representative 2D plasma map, obtained after albumin ad IgG removal, of a control subject. Arrows indicate the significant spots identified in the comparisons among the three groups and the zooms of some of the most significant ones. The bar graphs in Figure 2a,b show the intensity values of these spots for each of the three groups analyzed. The spots are grouped in three panels corresponding to the three most significant functions in which the proteins identified are involved: complement pathway and coagulation cascade, inflammation, and regulation of oxidative stress. Table 2 lists for each significant spot identified (p-value ≤ 0.05), the theoretical and observed point (pI), and molecular weight (MW), the MASCOT score, and the percentage of sequence coverage. The last three columns show the fold variation between mild or moderate versus the control group and between mild and moderate groups. Eighteen spots were differentially expressed between mild COPD patients and controls, in particular 7 spots were increased in the mild COPD group while 11 spots were decreased. Almost the same number of spots differed between the moderate and the control group (17 spots) although the identity for some of them was different. When comparing mild and moderate COPD groups, only five spots corresponding to four different proteins changed significantly. In particular, Protein AMBP, Ceruloplasmin, and Haptoglobin increased while the spot corresponding to Complement C4-B decreased.
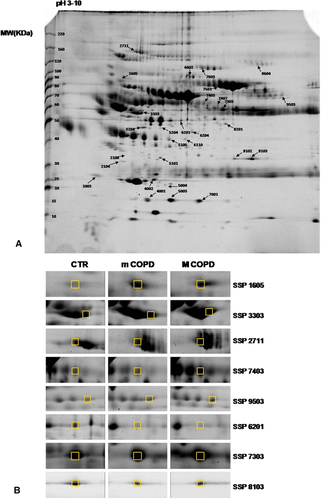
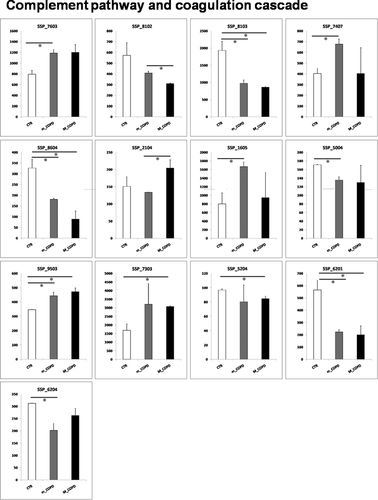

| N | SSP | Protein accession number | Protein identified | Theoretical (MW/pI) | Observed (MW/pI) | Score | Coverage [%] | (BPCO1-CTRL)/CTRL | (BPCO2-CTRL)/CTRL | (BPCO2-BPCO1)/BPCO1 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1003 | P01591 | Immunoglobulin J chain | 18/5.12 | 23/4.57 | 58 | 45 | −0.33 | ||
| 2 | 1605 | P00734 | Prothrombin | 70/5.64 | 93/4.94 | 72 | 39 | 1.09 | ||
| 3 | 2104 | P02760 | Protein AMBP | 39/5.95 | 33/4.98 | 67 | 33 | 0.53 | ||
| 4 | 2108 | P02647 | Apolipoprotein A-I | 30/5.56 | 37/5 | 155 | 56 | −0.82 | ||
| 5 | 2711 | P00450 | Ceruloplasmin | 122/5.24 | 164/5.18 | 135 | 38 | 3.14 | 1.36 | |
| 6 | 3204 | O43866 | CD5 antigen like | 38/5.28 | 50/5.32 | 190 | 69 | −0.30 | −0.25 | |
| 7 | 3303 | P02774 | Vitamin D-binding protein | 52/5.4 | 58/5.29 | 248 | 73 | 1.16 | ||
| 8 | 4001 | P00738 | Haptoglobin | 45/6.13 | 19/5.39 | 97 | 24 | 0.16 | 0.10 | |
| 9 | 4002 | P02647 | Apolipoprotein A-I | 30/5.56 | 22/5.49 | 272 | 86 | −0.10 | ||
| 10 | 5003 | P00738 | Haptoglobin | 45/6.13 | 19/5.69 | 66 | 26 | 0.30 | ||
| 11 | 5004 | P05452 | Tetranectin | 22/5.52 | 22/5.63 | 99 | 63 | −0.21 | ||
| 12 | 5101 | P02649 | Apolipoprotein E | 36/5.65 | 36/5.57 | 204 | 71 | −0.40 | ||
| 13 | 5105 | P00738 | Haptoglobin | 45/6.13 | 44/5.67 | 92 | 36 | −0.19 | 0.16 | 0.43 |
| 14 | 5204 | P02679 | Fibrinogen gamma chain | 51/5.37 | 49/5.57 | 119 | 51 | −0.12 | ||
| 15 | 6110 | O14791 | Apolipoprotein L1 | 43/5.6 | 44/5.85 | 58 | 34 | −0.31 | ||
| 16 | 6201 | P02679 | Fibrinogen gamma chain | 51/5.37 | 53/5.81 | 126 | 61 | −0.61 | −0.65 | |
| 17 | 6204 | P02679 | Fibrinogen gamma chain | 51/5.37 | 47/5.93 | 130 | 42 | −0.35 | ||
| 18 | 6602 | P06396 | Gelsolin | 85/5.9 | 101/5.91 | 141 | 39 | −0.19 | −0.61 | |
| 19 | 7001 | P00738 | Haptoglobin | 45/6.13 | 19/6.01 | 65 | 30 | 0.33 | ||
| 20 | 7303 | P02675 | Fibrinogen beta chain | 55/8.54 | 60/6.28 | 184 | 64 | 0.83 | ||
| 21 | 7403 | P01876 | Ig alpha-1 chain C region | 37/6.08 | 64/6.06 | 136 | 49 | 1.97 | ||
| 22 | 7407 | P02749 | Beta-2-glycoprotein 1 | 38/8.34 | 63/6.22 | 108 | 45 | 0.68 | ||
| 23 | 7501 | P02787 | Serotransferrin | 77/6.81 | 82/6.25 | 347 | 72 | 1.63 | ||
| 24 | 7603 | P00751 | Complement factor B | 85/6.67 | 120/6.05 | 129 | 36 | 0.49 | ||
| 25 | 8102 | P0C0L5 | Complement C4-B | 192/6.89 | 36/6.4 | 75 | 14 | −0.24 | ||
| 26 | 8103 | P0C0L5 | Complement C4-B | 192/6.89 | 35/6.54 | 80 | 19 | −0.49 | −0.55 | |
| 27 | 8201 | P02790 | Hemopexin | 51/6.55 | 52/6.31 | 71 | 39 | −0.25 | −0.19 | |
| 28 | 8604 | P00747 | Plasminogen | 90/7.04 | 128/6.66 | 151 | 39 | −0.45 | −0.73 | |
| 29 | 9503 | P02671 | Fibrinogen alpha chain | 94/5.7 | 70/7.65 | 167 | 32 | 0.28 | 0.36 |
3.3 Western Blot Validation
Western blot on pooled samples with specific antibodies against fibrinogen alpha chain and VDBP confirmed the OD difference between controls and the mild COPD group (Figure 3). The significance of the differences (p-value ≤ 0.05) was calculated by the Student t-test. The experiments were performed in triplicate. We decided to use technical replicates on pooled samples to validate our experimental data since plasma samples used for the 2-DE experiment were depleted with columns’ kit. So, using technical replicates has given us the chance to avoid the costly depletion of each of the 86 plasma samples even if we are aware of a loss in sensitivity.
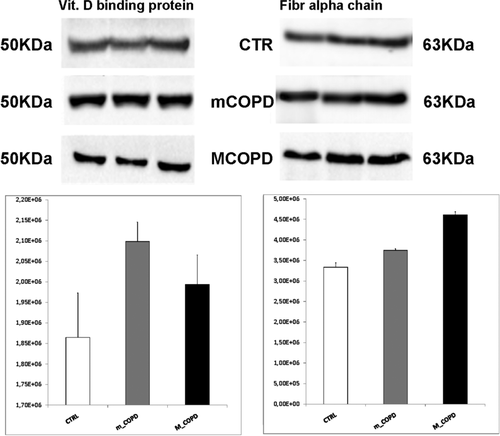
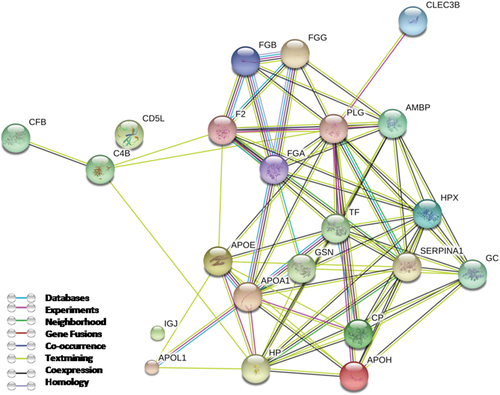
3.4 Protein–Protein Interaction Network Analysis
The protein network obtained with STRING database (Figure 4) shows how the majority of the proteins identified so far have a strong correlation in terms of known and predicted protein–protein interactions with the exception of two nodes that have not been connected to the network corresponding to CD5 antigen-like and immunoglobulin J chain.
4 Discussion
In this study, we applied a proteomic approach to identify potential biomarkers of early COPD by comparing data from mild and moderate COPD patients with an age- and sex-matched healthy control group. In the design of the experiment, the same proportion of current, never, and ex-smokers has been used. We decided to design the experiment in this way since our goal was to unravel protein biomarkers related to the COPD disease in the first stages using a sample population as similar as possible to reality with the majority of people being ex-smokers (65.5–78.6%). Current smokers are only 3 patients out of 43 in the control group and only 1 in each of the COPD groups, so current smokers would have little influence on the pooled samples. Having the same proportion in the three groups analyzed would have a normalization effect minimizing smoking influence on biomarkers and highlighting the ones that have little connection with smoking. Indeed, although it has long been recognized that smoking is the main risk factor not all smokers develop the disease and it is now recognized that never smokers may account for between one fourth and one third of all COPD cases.26-29 The factors determining the development of COPD are still poorly understood but many genetic and epigenetic factors, altered immune regulation, and abnormal repair mechanisms among others could be involved.30 Many scientific papers demonstrate that smoking cessation is able to reverse some inflammatory processes,31 so especially in the first stages of the disease ex-smokers would not have a great impact on the more relevant biomarkers identified in the present work. Most of the proteomic investigations performed so far on plasma of COPD patients have focused their attention on the more severe stages of the disease where the protein changes are supposed to be more relevant. Identification of proteomic signatures at the early stages of the disease would provide earlier and better diagnosis of patients. A total of 29 spots belonging to 20 unique proteins were identified using 2D-PAGE followed by MALDI-TOF mass spectrometer. Most between-group differences are proteins involved in regulating inflammation, complement and coagulation pathways, lipid metabolism, and oxidative stress. In the present work, three of the identified proteins are involved in the complement pathway. Among them, Complement factor B and Complement C4-B that show higher and lower expression values, respectively, in mild COPD versus the control group. The results of previous studies found lower serum levels of the inactive complement components C3 and C4 in patients with COPD compared with healthy subjects.32, 33 The third protein spot identified involved in the complement pathway is the one corresponding to apolipoprotein H (ApoH). ApoH, also known as beta-2-glycoprotein 1, regulates complement34 and is involved in the innate immune response; its plasma values decrease during the acute phase of infective inflammation.35 In this work, ApoH is higher in the mild group compared to the control group, but no difference was found between moderate and control group; this can suggest an initial involvement of the ApoH protein in trying to counteract the increasing inflammation status and a subsequent decrease when inflammation becomes higher. Proteases and antiproteases that regulate the coagulation process are also highlighted in the present work such as Protein AMBP and Prothrombin. Protein AMBP is an inter-alpha-trypsin inhibitor whose expression increased between the mild and the moderate COPD groups. In accordance with our findings, three peptides belonging to the AMBP protein have been identified so far as part of a panel of peptides patented as biomarkers of COPD whose values are reported to be increased.36 Prothrombin protein level was found elevated in mild compared to the control group, in accordance with Undas et al.37 Four of the spots identified in the present work are the ones belonging to the three fibrinogen chains. Many cross-sectional studies have found that the blood fibrinogen levels are higher in patients with COPD compared with healthy controls.38, 39 While all three fibrinogen chains are essential for the normal function of fibrinogen, due to their interconnected 3D structure, the gamma chain contains a number of sites that interact with other fibrin molecules, clotting factors, growth factors, and integrins.40 Fibrinogen interactions with integrins modulate the processes of platelet aggregation, inflammation, and wound healing.41 Moreover, fibrinogen gamma contains binding sites for different growth factors and cytokines that can serve as storage deposits to concentrate growth factors and cytokines in inflammatory and repair processes.42 We found a different behavior for spots belonging to the three fibrinogen chains, in particular we found an increase for some spots belonging to fibrinogen alpha and beta chains while there was a decrease in the fibrinogen spots belonging to the gamma chains in both mild and moderate COPD groups. The reason for this decrease is unclear, though some authors reported a decrease in some spots belonging to the gamma fibrinogen chain in plasma of early asthmatic patients in responses to allergen inhalation.42 The observed decrease was explained by the release of mediators from mast cells such as heparin and proteases that may activate fibrinogen gamma in sensitized asthmatic patients. Fourteen out of the 20 spots identified are proteins involved in regulation of response to stress in accordance with the increased oxidative burden demonstrated in the lungs and blood of patients with COPD.43-46 Among them serotransferrin, which is involved in the antioxidant defense mechanisms, is found to increase in both mild and moderate COPD groups. Higher serotransferrin expression have already been found in COPD lungs compared to control lungs. Philippot et al. suggest that an active iron sequestration by alveolar macrophages in COPD lungs could represent a protective mechanism to control and preserve the effects of free iron such as the formation of the highly toxic hydroxyl radical.47 However, besides having antioxidative properties, serotransferrin is a negative acute phase protein. Therefore, its levels can also be influenced by the systemic inflammatory state in COPD patients. This work also reveals the involvement of proteins related to the lipoproteins/lipids metabolism with antioxidant properties such as apolipoprotein L1, apolipoprotein A-I, and apolipoprotein E. Reduced levels of apolipoprotein A-I have been observed in the lungs of patients with COPD, compared with control smokers21 in line with the reduced levels of this protein observed in this work. This can be due to a deficient innate defense system in airway disease that could explain the increased susceptibility to infectious exacerbations.21 In the present work some acute phase proteins are highlighted. To date, it is not clear whether systemic inflammation is the cause or the consequence of the pathology. The acute phase proteins that emerge from this work beyond fibrinogen are haptoglobin, immunoglobulin alpha-1 chain C region, hemopexin, ceruloplasmin, and VDBP. A significant increase in three spots corresponding to haptoglobin are highlighted in this work. Haptoglobin has been shown to exert additional antioxidative, anti-inflammatory, and immunoregulatory effects by suppressing cellular immune responses and restoring homeostasis after inflammatory processes.48 Another interesting finding in our work is the detection of an higher expression of VDBP spot in the mild COPD group compared to the control one and a trend for an increase in the moderate group compared to the control group. In contrast to the direct relationship of vitamin D levels with lung function, VDBP levels have been reported to be inversely related to FEV1 in patients with alpha-1 anti-trypsin deficiency.49 VDBP has also been implicated as a candidate in the pathogenesis of COPD due to its role in macrophage activation and neutrophil chemotaxis.50 Since the transcription of the VDBP gene is affected by proinflammatory cytokines, VDBP might be considered an acute phase reactant.51 In our work of the three spots visualized in the gel corresponding to three isoforms, the one that shows significant changes in the mild group is the more basic spot. Looking at the intensity values of the other two spots we found that only two of the three isoforms show the same pattern of expression, while the central one seems to have an opposite trend showing lower intensity in the mild group even if not detected as significant (data not shown). The three VDBP spots might differ for their glycosylation status. VDBP occurs in several isoforms with various levels of glycosylation; glycosylation degree seems to regulate the inflammatory and immunological activities of VDBP.52 These findings suggest that the prevalence of one of the isoforms, rather than the total VDBP protein, is involved in the pathogenesis of COPD. Further glycoproteomics experiments will be needed to support this hypothesis. Another acute phase protein identified in this work is Ceruloplasmin which is higher in both mild and moderate COPD groups. These results are in agreement with higher serum ceruloplasmin concentrations previously observed in smokers and patients with COPD.53, 54 Higher levels of ceruloplasmin could be the result of both systemic oxidative stress and inflammation present in patients with COPD. In a recent paper, Milevoj et al. assessed the relationships of ceruloplasmin and other proteins to both oxidative stress and inflammation and speculate that in their cohort subjects ceruloplasmin concentration was mainly influenced by the presence of systemic inflammation.55
In conclusion, this work underlines how in the first stages of the disease it is possible to evaluate changes in plasma proteins involved in inflammation, regulation of blood coagulation, lipid metabolism, and oxidative stress, all of which have a strong correlation in terms of known and predicted protein–protein interactions, as revealed by the protein network obtained with STRING. The majority of the proteins identified related to inflammation tend to increase apart from the different behavior of the fibrinogen gamma chain, and a concomitant depletion of proteins with antioxidant properties can be found. A particular behavior is detected for VDBP, ApoH, and prothrombin which is higher in mild COPD patients and lower in the moderate COPD group. A possible explanation could be that the increase of these proteins in response to increased inflammation and oxidative stress burden is a temporary phenomenon that becomes overwhelmed by chronic exposure of airway and lungs to oxidative stress and inflammation. It would be interesting to monitor with the same approach all the GOLD stages of the COPD disease in order to see if the protein changes detected in this work are linked with disease severity or if they are distinctive of the early stages of the disease. Clinically useful biomarkers would provide earlier and better diagnosis of patients, allowing for an early intervention, thus possibly preventing the progression of COPD and its complications and comorbidities.
Abbreviations
-
- ACTH
-
- adrenocorticotropic hormone
-
- BALF
-
- bronchoalveolar lavage fluid
-
- COPD
-
- chronic obstructive pulmonary disease
-
- FEV1
-
- forced expiratory volume in 1 s
-
- FVC
-
- forced vital capacity
-
- GOLD
-
- global initiative for chronic obstructive pulmonary disease
-
- ICS
-
- inhaled corticosteroids
-
- LABA
-
- long-acting β-agonists
-
- LAMA
-
- long-acting muscarinic antagonist
-
- SABA
-
- short-acting β-agonists
-
- VDBP
-
- vitamin D binding protein
Acknowledgements
This study was supported by the “Fondazione Banco di Sardegna—Sassari—Italy” and by the “Ministero dell'Università e della Ricerca” Italy. A Visiting Professorship granted to Prof. Mangoni by the Department of Biomedical Sciences, University of Sassari (Italy), facilitated this work.
Conflict of Interest
The authors have declared no conflict of interest.




