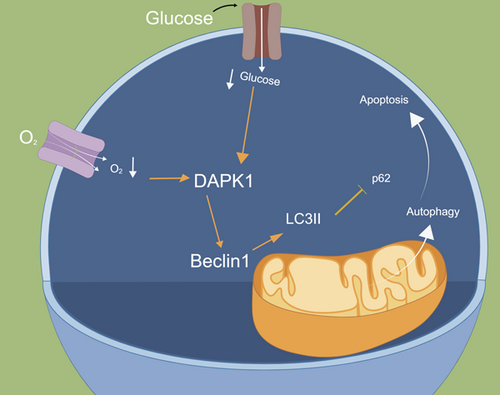
Overexpression of DAPK1 and Beclin1 under oxygen and glucose deprivation conditions promotes excessive autophagy and apoptosis in A549 cells
Linlin Wu and Wenxue Sun contributed equally to this study.
Abstract
In this study, we aimed to determine the specific roles of death-associated protein kinase 1 (DAPK1) and Beclin1 in non-small cell lung cancer (NSCLC) under oxygen and glucose deprivation (OGD). We found that OGD caused most cells to shrink, aggregate, and produce many vacuoles in the cytoplasm. Transmission electron microscopy revealed the presence of autophagic vesicles in the OGD group but not in the Control group. Moreover, the cell counting kit-8 assay showed that cell proliferation was reduced in the OGD group. Quantitative reverse transcription-polymerase chain reaction, western blot, and cell function assays showed that DAPK1 overexpression under OGD promoted apoptosis and autophagy in A549 cells. The coimmunoprecipitation assay confirmed the interaction between DAPK1 and Beclin1 protein. Moreover, knockdown of Beclin1 inhibited autophagy, but its overexpression promoted apoptosis in A549 cells. In vivo tumorigenesis experiment revealed that overexpression of DAPK1 promoted A549 cell apoptosis. Collectively, overexpression of DAPK1 and Beclin1 under OGD promoted excessive autophagy and apoptosis in A549 cells. Our study may provide a novel therapeutic target and theoretical basis for NSCLC treatment.
Graphical Abstract
Overexpression of death-associated protein kinase 1 (DAPK1) under oxygen and glucose deprivation (OGD) promoted A549 cell apoptosis and autophagy. Moreover, knockdown of Beclin1 inhibited autophagy. While overexpression of Beclin1 promoted A549 cell apoptosis. Collectively, overexpression of DAPK1 and upregulation of Beclin1 under OGD promoted excessive autophagy and aggravated apoptosis in A549 cells.
1 INTRODUCTION
Lung cancer is a major cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for 80%–85% of all lung cancers, with most patients in the advanced and inoperable stage.1 Chemotherapy is the primary treatment for NSCLC, but it has significant side effects.2 In recent years, the rapid development of molecular biology techniques has facilitated the recognition of NSCLC driver genes.3 Inhibitors and antibodies targeting driver gene-mediated pathways to inhibit tumor progression may prolong the patient survival and improve their quality of life.4 Due to its poor prognosis and resistance to radiotherapy and chemotherapy, the underlying molecular mechanisms need to be studied further to develop novel effective therapies for NSCLC.
Autophagy and apoptosis cause protein and organelle degradation, and cell stress leads to cell death; these processes play crucial roles in the pathophysiology of NSCLC.5 Autophagy and apoptosis occur when cells are under pressure.6 Autophagy is a cell protection mechanism against damaged organelles or protein aggregate degradation.7 It is a proteolytic process associated with NSCLC.8 Autophagy of small-molecule modulators shows great potential as anticancer drugs for cancer treatment.9 Therefore, regulation of autophagy has been suggested to improve the anticancer effects of chemotherapeutic drugs.10 Apoptosis is an evolutionarily conserved form of programmed cell death, which is essential for animal development and tissue homeostasis. Apoptosis plays a defensive role in immune response and cell destruction.11 Recent studies have shown that autophagy and apoptosis have the same effector and regulatory factors under the same conditions. Specifically, autophagy and apoptosis can be induced simultaneously, sequentially, independently, synergistically, or antagonistically when the cells are exposed to chemotherapeutic agents.12 However, the specific mechanisms underlying autophagy and apoptosis in NSCLC remain unclear.
Death-associated protein kinase 1 (DAPK1), a positive mediator of apoptosis, is a novel 160 kDa calmodulin-dependent serine/threonine kinase.13 Loss of DAPK1 activity may be an independent factor affecting the survival of patients with NSCLC. DAPK1 overexpression induces apoptosis and inhibits tumor cell metastasis.14 Guo et al.15 reported that extracellular vesicle-encapsulated microRNA-425-derived from drug-resistant cells facilitated NSCLC progression via the DAPK1-mediated phosphoinositide 3-kinase/protein kinase B pathway. As a key molecule controlling the autophagy pathway, Beclin1 can activate cell survival and death pathways.14 Beclin1 has been reported to successfully induce apoptosis in the target organs.16 Beclin1 loss and NF-E2-related factor 2 overexpression are associated with low survival rates in patients with NSCLC.17 Patients with NSCLC and low Beclin1 expression have been reported to show a more advanced stage of the disease with more lymph node metastasis and low tumor differentiation.18 However, the effects of DAPK1 and Beclin1 in NSCLC remain unknown. Therefore, we aimed to explore the specific roles of DAPK1 and Beclin1 in NSCLC herein.
In this article, we investigated the roles of DAPK1 and Beclin1 in NSCLC via in vitro and in vivo tumorigenesis experiments. Overexpression of DAPK1 and Beclin1 under oxygen and glucose deprivation (OGD) conditions were found to promote excessive autophagy and apoptosis in NSCLC cells. This study improves our understanding of NSCLC pathogenesis and provides new targets for NSCLC treatment.
2 RESULTS
2.1 A549 cells underwent autophagy under OGD conditions
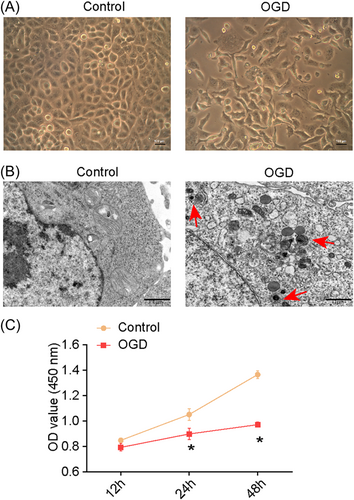
Most A549 cells shrank and aggregated under OGD conditions, showing many vacuoles in the cytoplasm (Figure 1A). Transmission electron microscopy (TEM) revealed the presence of autophagic vesicles in the OGD group (Figure 1B). The cell counting kit-8 (CCK-8) assay revealed the inhibition of cell proliferation in the OGD group (Figure 1C). Taken together, A549 cells initiated a defense mechanism and induced autophagy under OGD conditions.
2.2 Overexpression of DAPK1 under OGD conditions promoted apoptosis in A549 cells
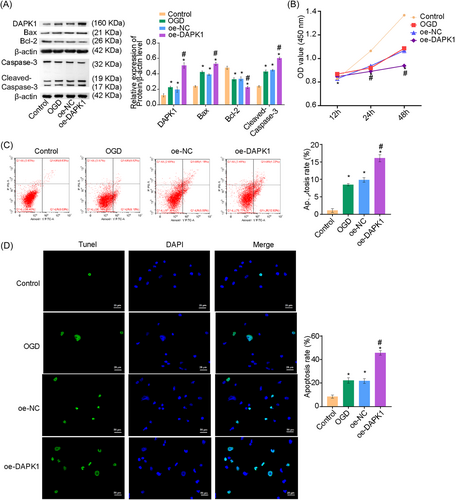
We hypothesized that DAPK1 may influence A549 cell apoptosis. To test this, we overexpressed (oe) DAPK1 in A549 cells. As shown in Figure 2A, DAPK1 expression was upregulated under OGD conditions. DAPK1 expression was higher in the oe-DAPK1 group than in the oe-negative control (NC) group, confirming the successful overexpression of DAPK1. Bcl-2-associated X (Bax) and cleaved-caspase-3 expressions were increased, and Bcl-2 expression was repressed under OGD conditions in the oe-DAPK1 group. CCK-8 assay results showed decreased cell proliferation in the oe-DAPK1 group (Figure 2B). Flow cytometry and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assays illuminated elevated apoptosis in the oe-DAPK1 group (Figure 2C,D). Collectively, overexpression of DAPK1 promoted apoptosis in A549 cells.
2.3 Overexpression of DAPK1 under OGD conditions promoted autophagy in A549 cells
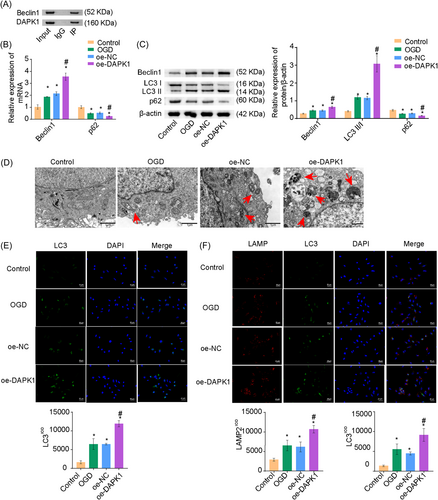
Various signaling pathways have been suggested to promote apoptosis by affecting autophagy in cancer cells.19 Here, we further verified the effects of DAPK1 on A549 cells. Coimmunoprecipitation (Co-IP) assay confirmed the interaction between DAPK1 and Beclin1 (Figure 3A). We then determined autophagy-related genes Beclin1, light chain 3-II/I (LC3II/I), and p62 levels. p62 levels were decreased, whereas Beclin1 and LC3II/I levels were elevated under OGD conditions in the oe-DAPK1 group (Figure 3B,C). TEM and monodansylcadaverine (MDC) staining revealed an increased number of autophagic vesicles and excessive autophagy in the oe-DAPK1 group (Figure 3D,E). Immunofluorescence (IF) revealed enhanced fluorescence of LC3 and lysosome-associated membrane protein 2 (LAMP2) in the oe-DAPK1 group, indicating increased autophagy (Figure 3F). Collectively, overexpression of DAPK1 promoted autophagy in A549 cells.
2.4 Knockdown of Beclin1 under OGD conditions inhibited autophagy in A549 cells
Next, we investigated whether Beclin1 affected autophagy in A549 cells. As shown in Figure 4A, Beclin1 expression was decreased in the si-Beclin1 group, indicating successful Beclin1 interference. LC3II/I levels were reduced and p62 levels were increased in the si-Beclin1 group (Figure 4B,C). TEM and MDC staining revealed decreased autophagy in the si-Beclin1 group (Figures 4D,E). These results suggested that Beclin1 knockdown inhibited autophagy in A549 cells.
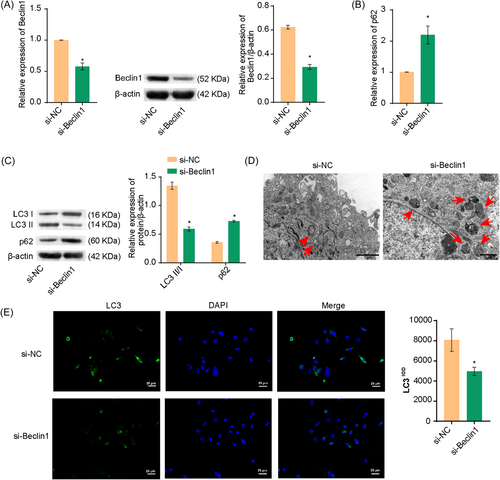
2.5 Knockdown of Beclin1 under OGD conditions reduced apoptosis in A549 cells
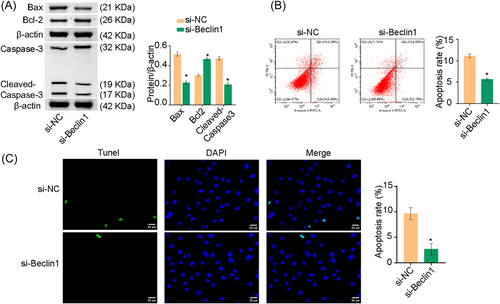
We found that Beclin1 affected autophagy in A549 cells. Next, we explored whether Beclin1 could also affect A549 cell apoptosis. Bax and cleaved-caspase-3 expression were decreased and Bcl-2 expression was elevated in the si-Beclin1 group than si-NC group (Figure 5A). Flow cytometry and TUNEL assays revealed that apoptosis was repressed in the si-Beclin1 group than in the si-NC group (Figure 5B,C). Thus, knockdown of Beclin1 under OGD conditions reduced apoptosis in A549 cells. Furthermore, Beclin1, Bax, and cleaved-caspase-3 expressions were increased and Bcl-2 expression was suppressed in the oe-Beclin1 group (Supporting Information: Figure S1A). Flow cytometry and TUNEL assays revealed increased apoptosis in the oe-Beclin1 group (Supporting Information: Figure S1B,C).
2.6 Overexpression of DAPK1 promoted A549 cell apoptosis in vivo
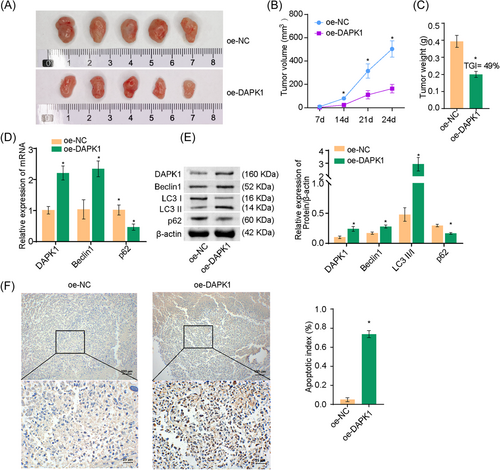
We previously demonstrated that the overexpression of DAPK1 under OGD conditions promoted apoptosis in A549 cells at the cellular level. Next, we tested whether the overexpression of DAPK1 promoted apoptosis in lung cancer cells in vivo. Figure 6A showed images of the tumor mass. The volume and weight of the tumors were suppressed in the oe-DAPK1 group than in the oe-NC group. The tumor growth inhibition (TGI) value was 49% in the oe-DAPK1 group compared with the oe-NC group (Figure 6B,C). Furthermore, DAPK1, Beclin1, and LC3II/I levels were promoted and p62 levels were suppressed in the oe-DAPK1 group (Figure 6D,E). TUNEL assay revealed increased apoptosis in the oe-DAPK1 group (Figure 6F). Tumor formation experiments in nude mice revealed that the overexpression of DAPK1 promoted A549 cell apoptosis.
3 DISCUSSION
Lung cancer is the most prevalent cancer worldwide, with NSCLC accounting for the majority of the lung cancer cases.20 Autophagy is a promising target for the development of cancer drugs.21 Autophagy is a highly conserved eukaryotic cell recycling process, the dysfunction of which leads to many disease pathogenesis.22 Tang et al.23 revealed that glyciparinic acid induced cell-protective autophagy in NSCLC cells by activating the inositol-requiring enzyme 1α–c-Jun N-terminal kinase/c-Jun pathway. Lin et al.24 found that autophagy induced by a derivative of tetrandrine played a protective role in NSCLC cells and suppressed apoptosis mediated by caspase-induced apoptosis. Moreover, the derivative of tetrandrine can induce apoptosis mediated by endoplasmic reticulum stress and promote autophagy in NSCLC cells. In this study, we used the OGD method to construct a cell model and confirmed the occurrence of autophagy in A549 cells under OGD conditions.
Apoptosis is the main cell death-regulating mechanism that is closely related to hypoxia-induced cell death.25 In tumors, the core cells are in a state of severe hypoxia and nutrient deprivation.26 OGD caused by insufficient blood circulation can reduce cancer cell survival and proliferation.27 DAPK1 exerts tumor suppressor functions and mediates multiple cellular processes, including apoptosis and autophagy.28 DAPK1 is a potential tumor suppressor gene, and its transcription is significantly reduced in lung cancer.27 DAPKs can be used as potential epigenetic biomarkers for early lung cancer detection.29 Li et al.30 reported that DAPK1 ameliorates inflammation, oxidative stress, and autophagy in lipopolysaccharide-induced acute lung injury via the p38 mitogen-activated protein kinase/nuclear factor-κB pathway. In this study, autophagy-related genes Beclin1 and LC3II/I expressions were increased, whereas those of p62 was decreased after the overexpression of DAPK1 under OGD conditions. In contrast, the levels of the apoptosis-related genes Bax and cleaved-caspase-3 were elevated, whereas Bcl-2 was repressed under OGD conditions. Our results revealed that the overexpression of DAPK1 under OGD conditions promoted autophagy and apoptosis in A549 cells. In vivo tumorigenesis experiment also confirmed that the overexpression of DAPK1 promoted apoptosis in A549 cells.
Previous studies have reported downregulation of Beclin1, an autophagy-related gene, in NSCLC, which may be related to its occurrence and development.31 High Beclin1 expression predicts better clinicopathological status and prognosis in patients with NSCLC.32 Bupivacaine treatment has been reported to increase Beclin1 expression in NSCLC cells.33 Downregulation of Beclin1, an autophagy-related gene, promoted cell growth and inhibited apoptosis in A549 cells.14 In this study, Beclin1 knockdown led to a decrease in LC3II/I expression and increase in p62 expression. However, Beclin1 overexpression increased the levels of the apoptosis-related genes Bax and cleaved-caspase-3 and decreased Bcl-2 expression. Therefore, knockdown of Beclin1 inhibited autophagy and promoted apoptosis in A549 cells.
However, there are some limitations in this study. We found that overexpression of DAPK1 under OGD promoted apoptosis and autophagy in A549 cells. However, a further knockdown of DAPK1 is needed for further validation. Furthermore, whether DAPK1 interacts with other proteins that participate in the formation of autophagosomes is unclear. Besides, we cannot add the images of the bright field for all images taken by a fluorescence microscope at present. Due to time and funding constraints, we are not able to solve these problems well at present, and in the future, with sufficient time and funding, we will perform a knockdown of DAPK1 to study its function and add the images of the bright field for the images taken by a fluorescence microscope.
In conclusion, our results revealed that the overexpression of DAPK1 and Beclin1 expression under OGD conditions promoted excessive autophagy and apoptosis in NSCLC cells. These findings improve our understanding of the mechanism underlying NSCLC pathology and can be used to develop novel treatment strategies for this disease.
4 MATERIALS AND METHODS
4.1 Cell culture and treatment
NSCLC cell line, A549, was purchased from ScienCell (Zhongqiaoxinzhou Biotech). Cells were cultured in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% double antibodies and divided into Control (A549 cells) and OGD groups. In the OGD group, A549 cells cultured with glucose-free Earle's balanced salt solution were placed in a hypoxia chamber for 4 h. Gas was continuously pumped with 95% N2 and 5% CO2 at 37°C (OM-14 Oxygen Monitor; SensorMedics Corporation) to maintain PO2 less than 1 mmHg for 48 h. Cells in the Control group were simultaneously cultured in glucose-free Earle's balanced salt solution in an anaerobic incubator for the same time. OGD state was changed by switching to normal culture conditions.34 To investigate its effects on A549 cells, we overexpressed DAPK1. Cells were divided into the Control, OGD, oe-NC (A549 cells transfected with oe-NC and then subjected to OGD), and oe-DAPK1 (A549 cells transfected with DAPK1 overexpression vector and then subjected to OGD) groups. To investigate whether autophagy and apoptosis in A549 cells were affected by Beclin1 expression, we knocked down and overexpressed Beclin1. Cells were divided into si-NC (A549 cells were transfected with si-NC and then subjected to OGD), si-Beclin1 (A549 cells were transfected with Beclin1 small interfering RNA [siRNA] and then subjected to OGD), oe-NC (A549 cells were transfected with si-NC and then subjected to OGD), and oe-Beclin1 (A549 cells were transfected with Beclin1 overexpression vector and then subjected to OGD) groups. oe-DAPK1, si-Beclin1, oe-Beclin1, and NC sequences were synthesized by HonorGene. Lipofectamine 3000 reagent (Invitrogen) was used to transfect the sequences into the cells.
4.2 TEM
A549 cells were digested and washed twice with phosphate-buffered saline (PBS). Then, A549 cells were centrifuged at 1500 rpm for 5 min, then the supernatant was discarded, and A549 cells were collected. Under an electron microscope, A549 cells were fixed with 2.5% glutaraldehyde, embedded, and stained with 1% OsO4. After alcohol dehydration, cells were stained with 3% lead citrate-uranyl acetate, and autophagosome formation was observed using JEM-1100 TEM (JEOL).
4.3 CCK-8 assay
A549 cells were seeded in a 96-well plate (5 × 103 cells/well, 100 μL/well). After culture, CCK-8 (10 μL, #NU679; Dojindo) was added for 12, 24, and 48 h. After incubation at 37°C and 5% CO2 for 4 h, absorbance at 450 nm was assessed using a Bio-Tek microplate (MB-530; Heals).
4.4 Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
DAPK1, Bax, Bcl-2, Caspase-3, and autophagy-related gene (Beclin1 and p62) expressions were determined via qRT-PCR. Briefly, Trizol was applied to extract the total RNA, and the complementary DNA (cDNA) reverse transcription kit (#CW2569; CWBIO) was performed to reverse transcribe the RNA into cDNA. Ultra SYBR Mixture (#CW2601; CWBIO) was utilized to determine the relative expression of genes with the ABI 7900 system. The reaction conditions were predenaturation at 95°C for 10 min, denaturation at 94°C for 15 s, and annealing at 60°C for 30 s, a total of 40 cycles. With β-actin as a reference gene, the 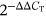 method was utilized to measure gene expression levels. The following primer sequences were used: H-DAPK1-F: GGGCGAGGGCTTCATTCTTC, H-DAPK1-R: CTTCCCAGACCATCACCACC; M-DAPK1-F: GACCTCCTACCCTGAGAGCA, M-DAPK1-R: GAGTCTCTCCGGGACACCA; H-Bax-F: TCACTGAAGCGACTGATGTCCC, H-Bax-R: ACTCCCGCCACAAAGATGGTC; H-Caspase-3-F: TGGCAACAGAATTTGAGTCCT, H-Caspase-3-R: ACCATCTTCTCACTTGGCAT; H-Bcl-2-F: AGCTGCACCTGACGCCCTT, H-Bcl-2-R: ACATCTCCCGGTTGACGCTCT; H-Beclin1-F: CATGGAGAACCTCAGCCGAA, H-Beclin1-R: ACAGCGTTTGTAGTTCTGACAC; M-Beclin1-F: GCTGTAGCCAGCCTCTGAAA, M-Beclin1-R: AATGGCTCCTGTGAGTTCCTG; H-p62-F: GCCATTGCGGAGCCTCATC, H-p62-R: TGTCAATTCCTCGTCACTGGA; M-p62-F: GGACCCATCTACAGAGGCTG, M-p62-R: ATCACAATGGTGGAGGGTGC; H-β-actin-F: ACCCTGAAGTACCCCATCGAG, H-β-actin-R: AGCACAGCCTGGATAGCAAC, M-β-actin-F: ACATCCGTAAAGACCTCTATGCC, M-β-actin-R: TACTCCTGCTTGCTGATCCAC.
method was utilized to measure gene expression levels. The following primer sequences were used: H-DAPK1-F: GGGCGAGGGCTTCATTCTTC, H-DAPK1-R: CTTCCCAGACCATCACCACC; M-DAPK1-F: GACCTCCTACCCTGAGAGCA, M-DAPK1-R: GAGTCTCTCCGGGACACCA; H-Bax-F: TCACTGAAGCGACTGATGTCCC, H-Bax-R: ACTCCCGCCACAAAGATGGTC; H-Caspase-3-F: TGGCAACAGAATTTGAGTCCT, H-Caspase-3-R: ACCATCTTCTCACTTGGCAT; H-Bcl-2-F: AGCTGCACCTGACGCCCTT, H-Bcl-2-R: ACATCTCCCGGTTGACGCTCT; H-Beclin1-F: CATGGAGAACCTCAGCCGAA, H-Beclin1-R: ACAGCGTTTGTAGTTCTGACAC; M-Beclin1-F: GCTGTAGCCAGCCTCTGAAA, M-Beclin1-R: AATGGCTCCTGTGAGTTCCTG; H-p62-F: GCCATTGCGGAGCCTCATC, H-p62-R: TGTCAATTCCTCGTCACTGGA; M-p62-F: GGACCCATCTACAGAGGCTG, M-p62-R: ATCACAATGGTGGAGGGTGC; H-β-actin-F: ACCCTGAAGTACCCCATCGAG, H-β-actin-R: AGCACAGCCTGGATAGCAAC, M-β-actin-F: ACATCCGTAAAGACCTCTATGCC, M-β-actin-R: TACTCCTGCTTGCTGATCCAC.
4.5 Western blot
Radioimmunoprecipitation assay lysis buffer (#P0013B; Beyotime Biotechnology) was used to extract the total protein from cells and tissues. Protein quantification was conducted on samples using a bicinchoninic acid protein assay kit, followed by adding sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer (#MB2479; MeilunBio). The mixture was then heated in boiling water for 5 min at 100°C. Proteins were adsorbed onto polyvinylidene fluoride membranes via gel electrophoresis. After incubation with primary antibodies against DAPK1 (#3008; 1:1000; CST), Bax (50599-2-Ig; 1:3000; Proteintech), caspase-3 (#9661; 1:1000, CST), Bcl-2 (12789-1-AP; 1:2000; Proteintech), Beclin1 (11306-1-AP; 1:1000; Proteintech), p62 (55274-1-AP; 1:600; Proteintech), LC3II/I (14600-1-AP; 1:2500; Proteintech), and β-actin (66008-1-Ig; 1:5000; Proteintech) at 4°C overnight, membranes were incubated with second antibodies anti-mouse (1:5000; SA00001-1; Proteintech) and anti-rabbit (1:6000; SA00001-2; Proteintech) for 90 min. β-Actin was used as an internal reference. and proteins were analyzed with Chemiscope6100 (CLINX).
4.6 Flow cytometry assay
A549 cells were digested with trypsin without ethylenediaminetetraacetic acid, centrifuged at 2000 rpm for 5 min, and approximately 3.2 × 105 cells were collected. After adding 500 μL annexin V-fluorescein isothiocyanate and 5 μL propidium iodide, A549 cells were reacted for 10 min at room temperature in the dark. Apoptotic cells were monitored via flow cytometry (A00-1-1102; Beckman).
4.7 TUNEL assay
TUNEL Apoptosis Detection Kit (#40306ES50; Yeasen) was utilized to assess apoptosis in A549 cells. Slices were fixed with 4% paraformaldehyde for 30 min, 100 μL proteinase K was added, and then slices were incubated at 37°C for 20 min. Slices were then immersed in PBS. Slices were dropped into 100 μL of 1× Equilibration Buffer for each sample to overlay the sample area completely and incubated for 10–30 min. Terminal deoxynucleotidyl transferase incubation buffer (50 μL) was added, and slices were incubated at 37°C for 1 h. After incubation, 4′,6-diamidino-2-phenylindole (DAPI) was added, slices were sealed with buffered glycerin, and observed under a fluorescence microscope.
TUNEL Apoptosis Detection Kit (KGA704; KeyGen Biotech) was used to detect apoptosis in tissue sections. Slices were incubated for 60 min at 60°C and dewaxed in water. One percent periodate was prepared, and slices were immersed in it and sealed for 12 min at room temperature. Proteinase K (100 μL) was dropped onto each sample and allowed to react for 20 min at 37°C. Then, the slices were immersed in PBS and later sealed with biotin (IH0125; Leagene Biotechnology) and labeled with horseradish peroxidase. Diaminobenzidine (ZLI-9018; ZSGB-BIO) was added for color development, and slices were sealed with buffered glycerin and observed under a light microscope.
4.8 Co-IP assay
To determine whether DAPK1 and Beclin1 interacted with each other, we performed Co-IP experiments. Cells were harvested for protein extraction, and equal amounts of protein extract were immunoprecipitated in lysis buffer with Beclin1 antibody at 4°C overnight. Then, protein A/G agarose beads were added to the IP mixture, incubated for 2 h, and washed thrice. The other procedures were the same as those used for western blot. Primary antibodies against DAPK1 (#3008; 1:1000; CST) and Beclin1 (11306-1-AP; 1:1000; Proteintech) were used for tests.
4.9 MDC staining
Autophagy was observed via MDC staining. First, A549 cells were cultured in a 6-well plate (3 × 105 cells/well) for 24 h, washed twice with PBS, and incubated in the dark with 0.05 mM MDC for 30 min at room temperature. A549 cells were then rinsed twice with PBS and observed under a fluorescence microscope.
4.10 IF
IF was used to analyze the colocalization of LC3 and LAMP2 in A549 cells. Slices were removed, fixed with 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100. After rinsing with PBS, 5% bovine serum albumin was added. Slices were incubated with properly diluted primary antibodies against LAMP2 (66301-Ig; 1:50; Proteintech) and LC3 (14600-1-AP; 1:50; Proteintech) overnight at 4°C, followed by incubation with goat anti-rabbit/mouse IgG (SA00013-2/SA00013-3; 1:200; Proteintech) antibodies at 37°C for 90 min. Slices were stained with DAPI and observed under a fluorescence microscope.
4.11 In vivo tumorigenesis experiment
Four-week-old male BALB/c nude mice were ordered from Hunan SJA Laboratory Animal Co., Ltd and subcutaneously injected with A549 cells after 1 week of adaptive feeding. The volume of cells injected was 2 × 106 cells per nude mouse, the injection volume was 100 μL, and the injection location was the right axilla. They were randomly divided into the oe-NC (oe-NC was transfected into A549 cells and then subcutaneously injected) and oe-DAPK1 (oe-DAPK1 was transfected into A549 cells and then subcutaneously injected) groups, with five mice in each group. The tumor volumes and weights at 7, 10, 14, 17, 21, and 24 days were measured in nude mice. Animal experiments were performed in accordance with national guidelines with the approval of the ethics committee of Jining Medical University (JNMC-2020-DW-RM-008).
4.12 Statistical analysis
Measurement data was analyzed using the GraphPad Prism8.0 software with mean ± standard deviation as the measure of statistical significance. Student's t-test was used to compare the data between two groups, and one-way analysis of variance was used to compare the data among multiple groups. Statistical significance was set at p < 0.05.
AUTHOR CONTRIBUTIONS
Linlin Wu: Conceptualization (equal); investigation (equal); methodology (equal); resources (equal); writing—original draft (equal). Wenxue Sun: Conceptualization (equal); investigation (equal); methodology (equal); software (equal); validation (equal); visualization (equal). Dehua Liao: Conceptualization (equal); investigation (equal); methodology (equal); resources (equal); software (equal); writing—original draft (equal). Yujin Guo: Formal analysis (equal); investigation (equal); software (equal); validation (equal). Qingying Si: Data curation (equal); investigation (equal); methodology (equal); software (equal); validation (equal). Dadi Xie: Data curation (equal); funding acquisition (equal); supervision (equal); validation (equal). Pei Jiang: Conceptualization (equal); funding acquisition (equal); methodology (equal); project administration (equal); writing—review and editing (equal). All authors have read and approved the article.
ACKNOWLEDGMENTS
The authors would like to thank the Department of Oncology, Tengzhou Central People's Hospital, Jining First People's Hospital, Jining Medical University, and Hunan Cancer Hospital, Central South University. We thank Figdraw (https://www.figdraw.com) for assistance in the graphical abstract drawing. This work was supported by the Natural Science Foundation of China (82272253 and 81602846) and Natural Science Foundation of Shandong Province (ZR2021MH145), the Taishan Scholar Project of Shandong Province (tsqn201812159), the China International Medical Foundation (No. Z-2018-35-2002), and the Bethune Charitable Foundation (No. B-19-H-20200622).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Animal experiments were performed according to national guidelines with the approval of the ethics committee of Jining Medical University (approved by JNMC-2020-DW-RM-008).
Open Research
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon request.