A homozygous stop codon in HORMAD2 in a patient with recurrent digynic triploid miscarriage
Abstract
Background
Recurrent miscarriage (RM) affects 1% to 5% of couples trying to conceive. Despite extensive clinical and laboratory testing, half of the RM cases remain unexplained. We report the genetic analysis of a couple with eight miscarriages and the search for their potential genetic etiology.
Methods
Short tandem repeat (STR) markers, single nucleotide polymorphic (SNP) microarray, and human DNA methylation microarray were used to analyze the genotypes of two miscarriages. Exomes sequencing was performed on DNA from the two partners and identified variants were validated by Sanger sequencing.
Results
STR marker genotyping demonstrated that the two available miscarriages are triploid digynic and resulted from the failure of Meiosis II. SNP microarray analysis revealed an additional Meiosis I abnormality that is the segregation of the two maternal homologous chromosomes in one triploid miscarriage. Whole-exome sequencing on DNA from the two partners identified candidate variants only in the female partner in two genes with roles in female reproduction, a missense in EIF4ENIF1 (OMIM 607445) and a stop gain in HORMAD2 (OMIM 618842). EIF4ENIF1 is a eukaryotic translation initiation factor 4E nuclear import factor required for the oocyte germinal vesicle breakdown, and HORMAD2 is part of the synaptonemal complex that was hypothesized to act as a checkpoint mechanism to eliminate oocytes with asynapsis during meiotic prophase I in mice.
Conclusion
While both genes may contribute to the phenotype, the Meiosis I abnormalities in the conceptions favor the causal role of HORMAD2 in the etiology of RM in this couple. This report illustrates the importance of comprehensively analyzing the products of conception to guide the search for the genetic causation of RM.
1 INTRODUCTION
Approximately 1% to 5% of couples trying to conceive experience recurrent miscarriage (RM), which is defined by the occurrence of at least two miscarriages before 22–24 weeks of gestation (Bender Atik et al., 2018). RM can be caused by or associated with a multitude of factors that include karyotype abnormalities in either of the two partners or in their conceptions, female factors such as thrombophilia, immunological, metabolic and endocrinological, and anatomical, or male factors (Colley et al., 2019; El Hachem et al., 2017; Stephenson & Kutteh, 2007). Consequently, the evaluation of couples with RM requires comprehensive clinical and laboratory investigations of both partners and of their miscarriages that are not always available in all medical centers. Another limitation of comprehensive investigations is the coverage of the cost of these laboratory tests by public and/or private medical insurance, which is not the same in all countries and health care systems. Therefore, the heterogeneity of RM in addition to the challenges associated with the implementation of standardized, comprehensive, and systematic evaluation of couples with RM have hampered the homogenization of this entity to facilitate its studies and reaching robust conclusions on its causative factors besides association studies on small cohorts of patients (Rull et al., 2012).
In humans, recurrent reproductive failure manifests mainly in three forms, infertility, recurrent molar pregnancy, and RM. The advent of next-generation sequencing in the past 15 years has greatly advanced the genetics of infertility and recurrent molar pregnancy and led to the identification of approximately 25 of their causative genes (Biswas et al., 2021; Colley et al., 2019; Robbins et al., 2019; Sang et al., 2021). However, few genes responsible for RM have been identified.
The goal of this study was to investigate the potential genetic cause of eight consecutive first trimester miscarriages over a period of 7 years and with no live birth in a couple of Indian origin that have remained unexplained despite extensive clinical and laboratory evaluations. Here we describe the analysis of their available products of conception (POCs) and their constitutive DNA whole exome sequencing (WES).
2 MATERIALS AND METHODS
2.1 Ethical compliance
This study involves human participants and was approved by McGill University Faculty of Medicine and Health Sciences Institutional Review Board (A01-M07-03A) in 2003 and renewed yearly since. Participants gave written informed consent to participate in the study.
2.2 Histopathological characterization of one product of conception
Archived formalin-fixed paraffin-embedded (FFPE) tissues were available only from the third product of conception (POC3). This POC was sectioned, and the sections were stained with hematoxylin and eosin and examined using bright field microscopy.
2.3 DNA extraction
Genomic DNA was extracted from peripheral blood samples from the couple and three family members using the Flexigene DNA Kit (QIAGEN) according to manufacturer's instructions. DNA extracted from freshly dissected chorionic villi was available from the seventh (POC7) and eighth POC (POC8). We attempted to genotype DNA extracted from the chorionic villi of POC3 using DNA FFPE Tissue Kit (QIAGEN). However, the DNA quality was not sufficient to obtain conclusive result.
2.4 SNP microarray
Affymetrix CytoScan HD microarray 750 K was performed at The Center for Applied Genomics (Toronto, Canada) on DNA from the male and female partners and from two of their miscarriages, POC7 and POC8, from which good quality DNA extracted from freshly dissected chorionic villi was available. The genotypes were assigned based on beta allele frequency (BAF) and visualized using Chromosome Analysis Suite 4.3 (ChAS 4.3). Assignment of the genotype from BAF was performed as previously described by applying the Mendelian inheritance scheme (Usui et al., 2019; Wirtenberger et al., 2005), designating the allele contributed by the partner (AA or BB), and determining the allelic contribution of the patient at informative loci where the patient has two different alleles (AB). The BAF analysis was then manually performed using Microsoft Excel 2010.
2.5 Methylation analysis
Genome-wide DNA methylation (DNAm) profiling on DNA from POC7 and POC8 and three control POCs was performed at the Center for Applied Genomics (TCAG; SickKids Research Institute, Toronto, Ontario, Canada). DNA was sodium bisulfite converted using the EpiTect Bisulfite Kit (EpiTect PLUS Bisulfite Kit, QIAGEN, Valencia, CA) according to the manufacturer's protocol. Modified genomic DNA was then processed and analyzed on the Infinium Human MethylationEPIC BeadChip (Illumina Inc, San Diego, California) according to the manufacturer's protocol. For quality control and normalization, the raw IDAT files were converted into β-values, which represent DNAm levels as a percentage (between 0 and 1) using the minfi Bioconductor package in R. Data preprocessing included filtering out nonspecific probes, probes with detection p-value >0.05 in more than 25% of the samples, probes located near single nucleotide polymorphic sites (SNPs) with minor allele frequencies above 1%, and X and Y chromosome probes. A total of 91,599 probes were removed, and a total of n = 774,260 probes were included in the methylation analysis as previously described (Choufani et al., 2020). Standard quality control metrics in minfi were used, including median intensity QC plots, density plots, and control probe plots. All samples passed quality control and were included in the analysis.
2.6 Genetic analysis
WES was performed on blood DNA from the patient and her partner at the Centre d'expertise et de services Génome Québec (Montreal, Quebec). Agilent SureSelect Human Exome library preparation was used for exome capture and Illumina NovaSeq 6000 PE100 for sequencing with 100x average coverage. Sequences were aligned to the human genome (GRCh37/hg19). The identified variants were validated and segregated in available family members by PCR amplification of genomic DNA and Sanger sequencing. The primers (Table S1) were designed using Primer3Plus (https://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi; Untergasser et al., 2007) and the UCSC reference genome (GRCh37/hg19; Kent et al., 2002). Variant pathogenicity prediction was performed according to ACMG guidelines (Richards et al., 2015) using VarSome (https://varsome.com/; Kopanos et al., 2019). NCBI Reference Sequence for HORMAD2 is NM_152510.4 and for EIF4ENIF1 is NM_019843.4. All variants detected have been submitted to the Leiden Open Variation Database (https://databases.lovd.nl/shared/individuals/00440109; patient ID 00440109). Simplex and multiplex short tandem repeat (STR) genotyping was performed on POC7 and POC8 as previously described (Khawajkie et al., 2017; Rezaei et al., 2022).
3 RESULTS
3.1 Patient and clinical history
The couple was referred to our laboratory by the Department of Clinical Genetics & Genetic Counseling, Mediscan Systems, Chennai, India, to investigate a possible genetic cause that could underlie their eight RM (all around gestational age of 8–9 weeks) over 7 years and with no live birth. The two partners are not smokers and are in good health. Their body mass indexes were slightly higher than normal ranges (26.7 and 25.7 for the female and male, respectively). They both have normal karyotypes using conventional cytogenetic analysis. Semen analysis of the male partner revealed mild asthenoteratozoospermia and hypospermia, but these abnormalities were not convincing to be the cause of their eight miscarriages. The male partner has one sister who had two children, and his parents did not have miscarriages or problems conceiving. The female partner had regular menstrual cycles. Her pelvic ultrasound revealed normal cavity, uterus size, and endometrium. In her mid-30s, her ovarian volumes were normal (8.99 cc and ovary 9.57 cc for the right and left ovaries, respectively) as well as her both adnexa. Antral follicular counts were 8 and 5 for the right and left ovaries, respectively, which is within normal range (4–24; Coelho Neto et al., 2018). The anti-Müllerian hormone (AMH), a hormone secreted by granulosa cells of maturing follicles, was measured also in her mid-30s and was normal (2.3 ng/mL). During her obstetrical evaluation, the patient tested only once positive for lupus anticoagulant. This test was repeated 3 months later and was negative. Nevertheless, the patient was given prophylactic treatment and put on Ecospirin 75 mg and low-molecular-weight heparin (Enoxaparin 40 mg/day) once a day from the start of the pregnancy until the date of the miscarriage for the last two miscarriages. As part of the patient work up, molecular karyotyping was performed on her seventh miscarriage (diagnosed as miscarriage by ultrasonography and microscopic histological evaluation) by quantitative fluorescent PCR analysis with primers from chromosomes 13, 18, 21, and X and Y, which demonstrated trisomy for all analyzed chromosomes suggesting a triploidy. The mother of the patient had three live births, one elective termination of pregnancy, and no history of miscarriages or primary or secondary infertility. The patient's mother had her menopause at the age of 55 years. The parents of the patient are fourth degree cousins. Also, there was no history of any form of reproductive failure in the maternal- or paternal grand-parents.
In conclusion, despite extensive testing, none of the above abnormalities in the two couples was convincing to explain their eight consecutive miscarriages with the first being when the female partner was in her mid-20s. The couple was then referred for genetic consultation with a suspicion of a maternal genetic defect, most likely because of the triploid miscarriage.
3.2 Histopathology and genotyping analysis of the miscarriages
Morphological evaluation of the third miscarriage, POC3, from which archived FFPE tissues were available, showed chorionic villi with some hydropic changes, intravillous fibrin, and absence of trophoblastic proliferation (Figure 1a). Fetal nucleated red blood cells were not present in the chorionic villi, and the available tissues did not contain any other embryonic or extra embryonic tissues. These findings are compatible with the diagnosis of an early arrested pregnancy. DNA extracted from freshly dissected tissues from POC7 and POC8 was used to determine the parental contribution to the POC genomes, using the PowerPlex 16 HS System (Promega, Corporation, Fitchburg, WI). This assay is a multiplex microsatellite genotyping kit that amplifies alleles at 16 markers from 15 chromosomes and the X and Y amelogenin gene (Table S2). The analysis of the DNA of POC7 and POC8, along with parental DNA, showed that at all the analyzed loci, the POCs received either two copies of the same maternal allele or two different maternal alleles along with a single paternal allele (Figure 1b). The amelogenin marker showed that the sex-chromosome complement of POC7 was XXY and POC8 was XXX. Hence, both POC7 and POC8 were triploid digynic. This suggested a defect in chromosome segregation during maternal meiosis.
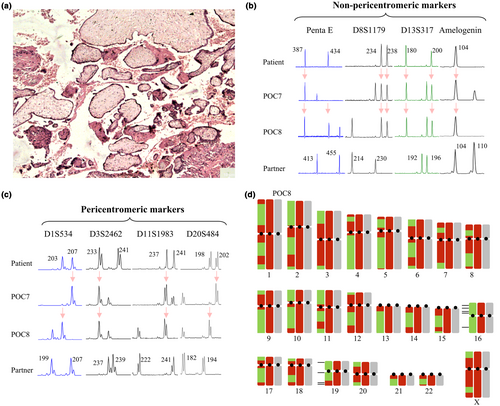
To evaluate whether these triploid digynic miscarriages resulted from the failure of Meiosis I (MI) or Meiosis II (MII), we investigated the segregation of alleles at 14 STR markers located in the vicinity of the centromeres. We identified six markers, from six different chromosomes, at which the mother is informative (heterozygous), and that were located at less than 7.4 Mb from the centromeres (Figure 1c; Table S2). Analysis of these markers showed the transmission of only one maternal allele to each of POC7 and POC8 with two exceptions, one in POC8 and one in POC7 at markers D10S1208 and D21S1436 located at 4.8 Mb and 7.4 Mb from the centromeres, respectively. These data suggested that the maternal triploidies are caused by the failure of the separation of sister chromatids at MII.
Altered number and distribution of crossovers have been associated with aneuploid miscarriages. To determine whether this is the case in the two triploid digynic conceptions of our patient, we performed SNP microarray analysis on DNA from POC7 and POC8, along with parental DNA. The analysis of the SNP microarray data on parental DNA confirmed their normal karyotypes without any detectable abnormality. The analysis of the two POCs confirmed their triploidies by the presence of four-allele peak tracks on all autosomes (AAA, AAB, ABB, and BBB; Figure S1). To map the positions of the maternal crossing overs, we filtered for positions where the father was homozygous, and looked at sites where the patient is informative and has two different alleles (Figure S2). Using this analysis, we found that a total of 60 crossing overs occurred in POC7 and 62 in POC8 (Figure 1d). These numbers when divided by two to correct for the two sets of maternal chromosomes in triploid digynic conceptions, give a total of 30 and 31 crossing overs, which is at the lower limit of the average number of crossing overs in female meiosis per haploid set of chromosomes, estimated to about 41.6 ± 11.3 (Lynn et al., 2004; Ottolini et al., 2015; Wirtenberger et al., 2005). Our SNP microarray analysis confirmed the failure of MII but revealed additional meiotic abnormalities in POC8. Judging by the allele type at the centromeric regions, this POC has received the two maternal homologous of chromosomes 16 and 19. On chromosome 19, we were able to observe one crossover event where maternal heterozygosity was reduced to homozygosity in POC8. For chromosome 16, we did not see a reduction to homozygosity, which implies either the absence of crossovers or the presence of the two reciprocal recombining chromatids which will appear as retained heterozygosity at all loci. These SNP microarray data were also confirmed using five and four informative STR markers from chromosomes 16 and 19.
Differentially methylated regions (DMRs) are associated with parent-of-origin specific transcription. In the female, these methylation marks are established during oocyte maturation, which takes place after the meiotic prophase I and before the completion of MII after fertilization. To investigate whether other meiotic abnormalities occurred in her two conceptions, POC7 and POC8, we assessed de novo DNA methylation at DMRs of imprinted regions using the Illumina Human Methylation-EPIC microarray. This analysis did not reveal statistically significant hypomethylation at maternally methylated DMRs (Figure S3) or genome-wide (within the limitation of the used methylation microarray) for the two POCs (Figure S4). Our data are in agreement with previous observations (Bourque et al., 2011) and showed a slightly higher level of DNA methylation in the two triploid digynic conceptions as compared to diploid biparental ones. This is due to the presence of DNA methylation on the two copies of the maternal genome, which represent 2/3 (67%) of the total copy numbers, whereas in a diploid biparental conception, maternal DNA methylation represents 1/2 (50%) of the total copy numbers (Bourque et al., 2011). This analysis expands the number of available methylation data on triploid digynic POCs that could be useful to interpret placental abnormalities in future studies.
3.3 Whole exome sequencing analysis
The maternal origin of two triploid miscarriages suggested a possible germline genetic defect in the patient at the origin of her RM. Because of the absence of any form of reproductive failure in the patient's parents and grand-parents, a recessive maternal defect was prioritized. We therefore performed WES on the patient's DNA. Identified variants were filtered for the following criteria: (1) homozygous, possible homozygous, or multiple heterozygous variants with minor allele frequencies less than 0.01 in gnomAD (v2.1.1 https://gnomad.broadinstitute.org/; Karczewski et al., 2020); (2) variants that are absent or very rare in 4400 in-house WES controls; (3) variants that correspond to stop gain, stop loss, invariant splice sites, frameshift insertion or deletion, or conserved missense variants with Combined Annotation Dependent Depletion (CADD) scores ≥10 (https://cadd.gs.washington.edu/; Rentzsch et al., 2021); and (4) variants in genes that are not highly mutated in in-house WES. Variants fulfilling these criteria were further investigated for their potential role in female reproduction (Figure S5). Two candidate genes met the above criteria and were validated by Sanger sequencing. Both variants were in the same run of homozygosity of 10 Mb on chromosome 22: a missense variant, c.554G>A, p.Arg185Gln in exon 5 of EIF4ENIF1 and a stop gain variant, c.505C>T, p.Gln169* in exon 9 of HORMAD2 (Figure 2). Furthermore, WES analysis of the male partner DNA and filtering his variants under the recessive mode did not reveal any variant that fulfill the above described criteria and that has a possible role in male reproduction (Figure S6).
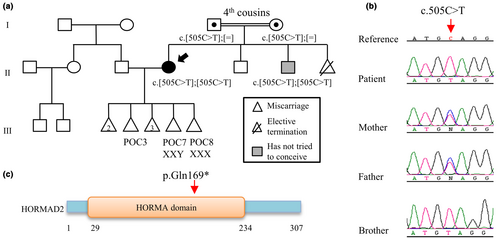
Analysis of the DNA from her two parents and one of her brothers showed that the parents are heterozygous carrier of the two variants in HORMAD2 and EIF4ENIF1, while the brother has the same genotype as the patient (Figure 2). However, the fertility status of the brother is unknown since he has not tried to conceive. The EIF4ENIF1 variant has an allele frequency of 0.0000159 in gnomAD Exomes (Karczewski et al., 2020) with 4 European non-Finnish males carrying this variant in heterozygous state and no reported homozygotes. This variant was predicted to be a variant of unknown significance (VUS) by the ACMG guidelines (Richards et al., 2015). The variant in HORMAD2 has an allele frequency in gnomAD Exomes (Karczewski et al., 2020) of 0.000556 with a total of 137 individuals carrying this variant in a heterozygous state and only two in a homozygous state (both South Asian males with no data about their reproduction). This variant frequency appears to be highest in South and East Asian populations and is predicted by the ACMG guidelines to be likely pathogenic (Richards et al., 2015). We next screened the exome data of 177 patients with RM but did not identify any patient with recessive variants in any of the two genes.
4 DISCUSSION
In this study, we report the genotypic analysis of two POCs from a female patient with eight RM and no live birth. Using simplex and multiplex STR genotyping, we demonstrated that both POCs were triploid digynic and have resulted from the failure of maternal MII. SNP microarray analysis confirmed the failure of maternal MII and revealed the presence of the two sister chromatids for all the chromosomes except for chromosomes 16 and 19, for which the two maternal homologs segregated in one of the two analyzed POCs. Analysis of DNA methylation confirmed that both POCs were triploid digynic, but did not reveal any additional DNA methylation abnormalities at imprinted DMRs.
WES revealed two homozygous variants, a missense VUS, p.Arg185Gln in EIF4ENIF1, and a likely pathogenic nonsense p.Gln169* in HORMAD2 segregating on the same haplotype from her two parents. We failed to identify a second patient with mutations in any of the two genes in a cohort of 177 patients with RM. This is not unexpected because RM is a highly heterogeneous entity, and from our experience, most are not caused by a recessive maternal genetic defect. EIF4ENIF1 codes for the eukaryotic translation initiation factor 4E nuclear import factor. In Drosophila, EIF4ENIF1 interacts with another protein to modulate ovarian development (Zappavigna et al., 2004). In mouse oocytes, knocking-down Eif4enif1 leads to failure of the oocyte nuclear envelop breakdown and oocyte development arrest (Pfender et al., 2015). To date, five different single-heterozygous variants in EIF4ENIF1 segregating in an autosomal dominant manner have been found in patients with premature ovarian insufficiency (POI; França et al., 2020; Kasippillai et al., 2013; Shang et al., 2022; Zhao et al., 2019). Four of these variants are missenses predicted by the ACMG (Richards et al., 2015) to be benign/likely benign, and only one is protein-truncating predicted to be VUS and segregating with the phenotype in a large family. However, no functional studies have yet demonstrated that these monoallelic variants cause POI. The fact that neither our patient nor her mother have had any feature of POI (amenorrhea, small ovarian volume, low AMH, and low antral follicle count) and that the patient's mother had her menopause at the age of 55 years make it unlikely for the EIF4ENIF1 variant in our patient to be the cause of her RM.
HORMAD2 mutations have not been associated with reproductive failure in humans. However, there has been extensive research regarding HORMAD2 functions in knockout mouse models. HORMAD2 is part of the synaptonemal complex (SC), which is a proteinaceous structure present only in meiotic cells (Xie et al., 2022). The SC consists of two axial elements that form along the longitudinal axes of sister chromatid pairs and a central element. Sister chromatids are bound to the axial elements and to each other via cohesion complex. During prophase I, HORMAD2 localizes mainly to unsynapsed regions of the axial elements and has a supporting role in completing synapsis (Kogo et al., 2012). Additionally, HORMAD2 plays an essential role in efficient ATR recruitment to unsynapsed chromatin, H2AX phosphorylation, and meiotic silencing of unsynapsed chromatin (Kogo et al., 2012; Wojtasz et al., 2012). Hormad2−/− male mice are infertile because the lack of HORMAD2 in the presence of normal asynapsis between the largely nonhomologous X and Y chromosomes leads to spermatocyte apoptosis due to impaired meiotic sex chromosome inactivation. In females, asynapsis does not naturally occur on sex chromosomes, and consequently, HORMAD2 deficiency is tolerated, and the females are fertile and have normal litter sizes. The only abnormalities observed in their oocytes are a slight increase in the number of incomplete synapsis in fetal ovaries and a slight decrease in the frequency of chiasmata formation in in vitro matured metaphase I oocytes in adult mice, as compared to wildtype. Incomplete synapsis is well known to be associated with a lower than normal number of crossing overs, which is in agreement with the borderline low number of crossing overs observed in the two triploid POCs of our patient. Since HORMAD2 in mice is thought to be involved in the elimination of oocytes with asynapsis via a checkpoint mechanism, it is possible that its recessive mutation in our patient prevented her oocytes with synaptic errors from being eliminated. These oocytes progressed to MII were fertilized, but failed to extrude the second polar body. Alternatively, the abnormal segregation of the two homologous chromosomes 16 and 19 in POC8 may have been caused by the lack of normal HORMAD2 from the axial elements of the SC, which may have impaired sister chromatid attachment by cohesion complex and led to their precocious separation. This followed by the random segregation of their four chromatids at MI may have led to the presence of the two homologous chromosomes 16 and 19 in POC8 (Handyside et al., 2012; Ottolini et al., 2015). This suggestion is in line with the increase of univalents in Hormad2−/− oocytes (Wojtasz et al., 2012), which is well documented to promote the precocious separation of sister chromatids in MI (Capalbo et al., 2017). Also, the presence of two triploid conceptions in our patient is in agreement with the high prevalence of hyperploidy observed in embryos derived from null-female mice for HORMAD1, which co-localizes and interacts with HORMAD2 (Kim et al., 2014; Wojtasz et al., 2009, 2012).
Human and murine HORMAD2 proteins are highly conserved with 76% of identity and 87% of similarity. While Hormad2-deficient female mice are fertile and have normal litter sizes, our patient did not achieve any live birth, which suggests potential differences in how HORMAD2 functions in mice and humans. It is also possible that Hormad2-deficiency might have a more severe impact on female fertility in other mouse strains than in the strain studied by Wojtasz et al. (2012) and Kogo et al. (2012).
Although the male partner had mild abnormalities in the semen analysis, his karyotype by culture-based cytogenetic did not reveal any chromosomal abnormality, which was also confirmed by SNP microarray analysis. WES analysis on his DNA did not reveal any plausible causative candidate variant. The fact that both analyzed POCs are digynic triploid is in support of a maternal genetic defect underlying the eight consecutive miscarriages.
In summary, while we cannot exclude a possible contribution of the EIF4ENIF1 variant to the phenotype of the patient, the presence of MI and MII abnormalities in two of her conceptions are in favor of the causative role of HORMAD2 variant, which remains to be confirmed in more patients in future studies. Our study highlights the genetic complexity of RM and the importance of SNP microarray in determining the meiotic origin of triploid conceptions and guiding the search for their causative genes.
AUTHOR CONTRIBUTIONS
Manqi Liang and Rima Slim: Planning and conducting the project, data analysis, and drafting the manuscript. Beena Suresh and Sujatha Jagadeesh: Referring the patient, gathering clinical information and materials from the conceptions. Eric Bareke and Jacek Majewski: Processing raw data of exome sequencing and conducting variant calls. Sanaa Choufani and Rosanna Weksberg: performing the methylation microarray and guiding its analysis. All authors read and commented on the manuscript.
ACKNOWLEDGMENTS
We thank the couple and their family members for participating in our study, Susmitha Jaganathan for technical help, Dr. Anna Naumova for discussion and critical reading of the manuscript, and Dr. Trilochan Sahoo for the analysis of SNP microarray on blood DNA of the couple. We acknowledge the use of the Histopathology Core Facility of the Research Institute of the McGill University Health Centre and the Centre d'expertise et de services Génome Québec.
FUNDING INFORMATION
This study was supported by the Canadian Institute of Health Research to RS (PJT-180509). ML was supported by the Research Institute of the McGill University Health Centre Desjardins Studentship, McGill University Faculty of Medicine and Health Sciences Internal Studentship, and Travel Funding Support from the Réseau Québécois en Reproduction and the Department of Human Genetics of McGill University.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
ETHICS STATEMENT
This study involves human participants and was approved by McGill University Faculty of Medicine and Health Sciences (A01-M07-03A) in 2003 and renewed yearly since. Participants gave written informed consent to participate in the study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




