Precision therapy for a medically actionable ATP1A3 variant from a genomic medicine program in an underserved population
Texome Project Members are listed in supplemental table 3.
Abstract
Background
Genomic medicine is revolutionizing the diagnosis of rare diseases, but the implementation has not benefited underrepresented populations to the same degree. Here, we report the case of a 7-year-old boy with hypotonia, global developmental delay, strabismus, seizures, and previously suspected mitochondrial myopathy. This proband comes from an underrepresented minority and was denied exome sequencing by his public insurance.
Methods
After informed consent was obtained, buccal cells from the proband were collected and whole exome sequencing was performed. Illumina Dragen and Emedgene software was used to analyze the data at Baylor Genetics. The variants were further intepreted according to ACMG guidelines and the patient's phenotype.
Results
Through whole-exome sequencing (WES) under the Community Texome project, he was found to have a heterozygous de novo pathogenic variant in the ATP1A3 gene located on chromosome 19q13.
Conclusion
In retrospect, his symptomatology matches the known medical conditions associated with the ATP1A3 gene namely Alternating Hemiplegia of Childhood 2 (AHC), a rare autosomal dominant disorder with an incidence of 1 in one million. His single nucleotide variant, (c.2401G>A, p.D801N), is predicted to be damaging. The specific amino acid change p.D801N has been previously reported in ClinVar along with the allelic variant p.D801Y and both are considered pathogenic. The identification of this variant altered medical management for this patient as he was started on a calcium antagonist and has reported no further hemiplegic episodes. This case illustrates the value of implementing genomic medicine for precision therapy in underserved populations.
1 BACKGROUND
The ATP1A3 gene (OMIM:*182350) on chromosome 19q13 encodes for the α3 subunit of a transmembrane Na+/K+ ATPase. This electrogenic transmembrane ATPase is ubiquitously expressed in the central nervous system and was first described in 1957 (Capuano et al., 2020). The α3 subunit is expressed in GABAergic neurons in multiple brain areas, including several thalamic nuclei, the cortex, cerebellum, red nucleus, hippocampus, reticulotegmental nucleus of the pons, and all the nuclei of the basal ganglia. The α3 subunit allows for rapid normalization of the transmembrane gradient due to its lower affinity for Na+ and K+ and lower voltage dependence (Capuano et al., 2020). Pathogenic variants in ATP1A3 are associated with Alternating Hemiplegia of Childhood 2 (AHC, OMIM: #614820), Rapid Onset Dystonia Parkinsonism (RDP, OMIM: #128235), Developmental and epileptic encephalopathy 99 (OMIM #619606) and CAPOS Syndrome (OMIM: #601338) (cerebellar ataxia, areflexia, pes cavus, optic atrophy, and sensorineural hearing loss; Salles et al., 2021). Disease-causing variants related to Alternating Hemiplegia of Childhood 2 in ATP1A3 are most frequent in exons 17 and 18 (Capuano et al., 2020).
There are 3 variants that account for 60% of the individuals with the Alternating Hemiplegia of Childhood 2 phenotype. One variant, p.D801N, accounts for 30%–40% of cases while p.E815K and p.G947R account for 16%–35% and 8%–15% of cases, respectively (Heinzen et al., 2014). These small amino acid alterations lead to functional problems because these missense mutations change the tertiary structure of the protein (Rosewich et al., 2012). The incidence of AHC caused by ATP1A3 is about 1 in one million, and next-generation sequencing technology can assist in diagnosing patients with this condition. Indeed with more advanced sequencing, a new understanding of the genotype–phenotype relationships of ATP1A3-related syndromes has emerged (Panagiotakaki et al., 2015; Salles et al., 2021). While improved DNA sequencing technology has decreased the cost of whole exome sequencing and whole genome sequencing, leading to more routine use and increased diagnosis of rare disease, this benefit has still not been accessible to underrepresented minorities (Fraiman & Wojcik, 2021; Hartin et al., 2020). Patients with no insurance or underinsured individuals do not have full access to personalized genomic medicine in current conditions (Goldenberg et al., 2013). The Texome Project aims to close this disparity gap by providing personalized genomic medicine and longitudinal follow-up to underrepresented minorities in Texas who lack the financial means to obtain exome sequencing.
Here, we report the case of a 7-year-old boy with an undiagnosed, rare disease. The patient's public insurance denied coverage for whole exome sequencing and the family could not pay out-of-pocket to do the testing on their own. With the help of The Texome Project, this patient was able to have exome sequencing and receive a diagnosis for his suspected genetic condition.
2 CASE PRESENTATION
The proband is a 7-year-old boy born to non-consanguineous healthy parents. The mother reported an uneventful pregnancy. The patient was born at 34 weeks and delivered via C-section for oligohydramnios. After delivery, he had decreased movements and spent 6 days in the Neonatal Intensive Care Unit (NICU) due respiratory distress and feeding difficulties. He developed epilepsy at 1 month and respiratory insufficiency at 2 months. Testing of a muscle biopsy performed in Mexico at 8 months old suggested a mitochondrial myopathy per the proband's mother. Subsequent evaluations revealed an atrial septal defect that required a cardiac catheterization at 6 years. His development was also delayed. He began to roll at 12 months, sit alone at 24 months, and crawl at 4 years. Currently, he uses braces, a walker, or a wheelchair for ambulation. He began to speak single words at 3 years of age. He currently does not speak in full sentences but has about ten words in total. He is not toilet trained. Growth is <1 percentile for height and weight. He has recurrent respiratory infections, constipation, strabismus, and aggravated behavior when frustrated.
His physical exam is notable for brachycephaly, myopathic facies, flat midface, pointed chin, low posterior hairline, and fine dark hair on his chin, chest, and back. The proband also has hypertelorism, downward slanting palpebral fissures, epicanthal folds, prominent ears, small depressed nasal root, anteverted nares, short columella, rounded nose tip, long philtrum, tented lips, and high arched palate. His chest shows pectus excavatum. He has brachydactyly and diminished palmar creases in his hands with prominent fingertip pads and a proximally placed thumb. The feet show metatarsus adductus, plantar creases, and mild hallux valgus. Neurological exam revealed bilateral ptosis and mild bilateral clonus.
Prior testing was performed for a possible mitochondrial disorder based on his clinical features. However, the testing did not support this possibility. His testing included normal lactic acid, normal pyruvic acid, normal CMP besides a slightly elevated BUN, normal TSH/T4, mild CK elevation (201, normal 30–200), normal urine organic acids, nonspecific urine amino acid abnormalities, normal plasma acylcarnitines, and mild elevations of various plasma amino acids in a nonspecific pattern. A neuromuscular disorders panel revealed one heterozygous variant of uncertain significance (SGCB, exon 2, c.68G>A, p.Arg23His) in a gene associated with an autosomal recessive disorder. A Combined Mito Genome Plus Mito Focused Nuclear Gene Panel/Sequencing and Deletion Analysis, which encompassed 202 nuclear-encoded mitochondrial genes, showed two heterozygous variants of uncertain significance (DLD c.768G>A, p.R263H and NDUFS7 c.101T>C, p.V34A) present in genes associated with autosomal recessive disorders.
3 GENETIC ANALYSIS AND PRECISION MEDICINE IMPLICATIONS
Informed consent was obtained from the parents of this research participant under the Baylor College of Medicine Institutional Review Board Protocol H-49277 prior to participation in study activities. We performed whole-exome sequencing (WES) on the DNA isolated from cheek cells of the proband and his parents. The isolated DNA was indexed into separate libraries, which were enriched by in-solution exome capture probes, purified, and pooled for sequence analysis on an Illumina platform for paired-end read sequencing. The data were aligned to the human reference genome build GRCh37 (HG19) using the Illumina Dragen BioIT Platform. The Illumina Dragen haplotype variant calling system and Illumina Dragen genome-wide depth-based CNV caller with custom modifications from Baylor Genetics were used to call the variants from the samples. Emedgene, an automated genetic interpretation platform, was utilized for variant annotation and interpretation with proprietary algorithms and open-source data sets. The variants were interpreted according to ACMG guidelines and the patient's phenotype. Any synonymous variants, intronic variants not affecting splice sites, and common benign variants are not included in analyses unless previously reported as a possibly pathogenic variant (Richards et al., 2015). If the data quality of variants from WES was insufficient, the variants were confirmed via Sanger sequencing. Any variants with low likelihood of diagnosis were not reported in the proband report. The decision to include or not include variants in a possible diagnosis was based on the clinical information given.
WES detected a heterozygous missense variant in exon 17 of the ATP1A3 gene (NM_152296.5). This is a de novo variant as both of his patients are negative for the variant. The variant is in position chr19:42474557C>T (HG19); c.2401G>A, p.D801N. This nucleotide change leads to an amino acid change in position 801 where the negatively charged aspartic acid was substituted by asparagine, a polar uncharged amino acid (Table 1).
| Gene | Accession number | Variant location | Nucleotide change | Amino acid change | Mutation taster | CADD | REVEL | Zygosity | Variant class |
|---|---|---|---|---|---|---|---|---|---|
| ATP1A3 | NM_152296.5 | chr19:4247557 C>T | c.2401G>A | p.Asp801Asn | D | 29.400 | 0.905 | Het | Pathogenic |
- Abbreviation: D, disease causing.
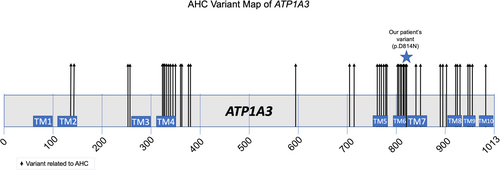
ATP1A3 encodes for Na+/K+ ATPases found in the central nervous system. The α subunit is a part of the catalytic subunit and has 4 isoforms with 3 cytoplasmic domains and 1 transmembrane domain in each (Capuano et al., 2020; Carecchio et al., 2018). The cytoplasmic domain consists of the phosphorylation domain, the nucleotide-binding domain, and the actuator domain. The transmembrane domain forms 10 transmembrane helices. AHC-causing variants typically affect ion binding sites or the transmembrane segments, while a smaller portion of them are found in the transmembrane helices leading to changes in ion binding and transport. The proband's change is localized to the area of TM6 and the intracellular loop between TM6 and TM7. (Figure 1). This variant was not present in gnomAD (Table 2) and is predicted to be damaging.
| Category | Expected SNVs | Observed SNVs | Constraint metrics | o/e |
|---|---|---|---|---|
| Synonymous | 274.1 | 302 | Z = −1.32 | 1.10 |
| Missense | 635.1 | 186 | Z = 6.33 | 0.29 |
| Loss of Function (LoF) | 47.7 | 0 | pLI = 1 | 0 |
In retrospect, it was noted that the patient was having episodic hemiplegia of the left side of the body. These episodes could be associated with GI symptoms and resolved completely with sleep. Flunarazine, which is a calcium antagonist has been used for several decades but is not approved for marketing in the United States. It can be obtained from other countries. The patient was started on Flunarazine, which he was prescribed, and the family was instructed how to obtain the drug in Mexico by their neurologist. The patient has had no further hemiplegic episodes to date. In this circumstance, the genetic information directly impacted the medical care of this patient.
4 DISCUSSION
ATP1A3-related disorders were originally characterized as a group of distinct syndromes, which are now thought to be on a clinical spectrum. There are three classical syndromes associated with ATP1A3: Alternating Hemiplegia of Childhood 2 or AHC (OMIM: #614820), Rapid Onset Dystonia Parkinsonism (RDP, OMIM: #128235), and CAPOS Syndrome (OMIM: #601338) or cerebellar ataxia, areflexia, pes cavus, optic atrophy, and sensorineural hearing loss. There has also been an increase in non-classical phenotypes associated with mutations in ATP1A3 in recent years (Salles et al., 2021).
AHC is a rare autosomal dominant disorder with the most common pathogenic variants being p.D801N, p.E815K, and p.G947R (Salles et al., 2021). In an effort to make the diagnosis of AHC easier, seven diagnostic criteria have been established: (1) onset of symptoms before 18 months of age; (2) repeated episodes of hemiplegia involving the right or left side of the body, at least in some episodes; (3) episodes of bilateral hemiplegia or quadriplegia, starting either as a generalization of a hemiplegic episode or bilaterally; (4) other paroxysmal disturbances including tonic/dystonic attacks, nystagmus, strabismus, dyspnea, and other autonomic phenomena occurring during hemiplegic attacks or in isolation; (5) immediate disappearance of all symptoms on going to sleep, with recurrence 10 to 20 min after awakening in long-lasting attacks; (6) evidence of developmental delay, learning disability, neurological abnormalities, choreoathetosis, dystonia, or ataxia; and (7) not attributable to another disorder. Although not all cases will satisfy these criteria, those that do are considered typical. More specifically, typical cases meet criteria 1, 2, 3, and 7 (Neville & Ninan, 2007). Epilepsy is also common in patients with AHC (Capuano et al., 2020).
Our patient meets diagnostic criteria 1, 2, 4, 5, 6, and 7 and is classified as having atypical AHC. The hemiplegic episodes were not a prominent part of his initial evaluation but were present (criteria 2). The proband suffered from hypotonia at the time of birth and had a muscle biopsy at 8 months of age in an effort to diagnose his decreased muscle movement and tone (criteria 1). He was diagnosed with strabismus (criteria 4) for which he is followed by ophthalmology. His dystonic movements seem to subside at night while resting according to his family (criteria 5). The proband has significant developmental delays. At the age of 7, the proband only speaks about 10 words. He does not speak in full sentences and is not toilet-trained or fully ambulatory (criteria 6). So far, his symptoms cannot be attributed to any other disorder (criteria 7). He also began having seizures at 1 month of age. Also of note, he has had previous genetic testing, imaging, and laboratory testing that were all non-diagnostic.
The proband was found to have one of the most common AHC mutations, D801N. This variant is documented in ClinVar as pathogenic from a number of sources (ClinVar Variation ID 37107). This variant was reported in early reports of ATP1A3 being linked to hemiplegic migraine (Heinzen et al., 2012; Rosewich et al., 2012). A mouse model of this variant has been studied and has phenotypes that overlap with AHC (Hunanyan et al., 2015). Some genotype–phenotype correlation studies support a less severe phenotype for this variant(Capuano et al., 2020; Panagiotakaki et al., 2015). Patients with the p.D801N change tend to have fewer hemiplegic attacks, less dystonia, and are able to ambulate, indicating a milder phenotype. Patients with the p.E815K variant have more severe cognitive disability, dystonia, and epilepsy (Panagiotakaki et al., 2015). With whole exome sequencing (WES), we were able to end the diagnostic odyssey for this family. Some studies have shown that symptoms of AHC can be improved with the use of Flunarizine (Cordani et al., 2021; Kusunoki et al., 2020). Flunarizine is a selective calcium channel entry blocker with calmodulin binding properties and histamine H1 blocking activity (Upadhyay & Ali, 2018). While this drug has mostly been used in Italy and Japan, these studies have shown reduction in symptom severity and frequency of hemiplegic attacks (Kusunoki et al., 2020). The ability to use genetic testing to find a medically actionable diagnosis with a known treatment is a clear-cut example of precision medicine at work and indeed Flunarazine was started with early signs of benefit for this case.
The Texome Project is a clinical study that provides whole exome sequencing for underserved populations and ethnic minorities in an effort to end the disparities in genomic medicine. It has been shown that whole exome sequencing and whole genome sequencing have improved the ability to detect rare variants and routinely achieve diagnostic rates of 25%–35% in pediatric cohorts (Hartin et al., 2020). In this case, there are some therapeutic implications for our patient given the diagnosis. This suggests that genomic sequencing in underserved individuals with suspected rare disease has potential impact that can be medically actionable. Access to such genomic technology should therefore be available to each and every patient.
In summary, the present case reports a pediatric patient with a heterozygous de novo ATP1A3 pathogenic variant leading to a diagnosis of Alternating Hemiplegia of Childhood 2. Through the Texome Project, the possibility of precision medicine was illustrated in an underserved community and progress was made in decreasing disparities in genomic medicine.
AUTHOR CONTRIBUTIONS
Carrie A. Schmid and Rebecca O. Littlejohn recruited the human subject and provided clinical evaluation. Liesbeth Vossaert performed and interpreted the exome sequencing described, Cara P. Ford, Ryan German and Blake Vuocolo wrote the first draft of the manuscript. Michael Wangler and Carrie A. Schmid oversaw the project. All authors edited and contributed to the final draft.
ACKNOWLEDGMENTS
This study was funded by a grant from NHGRI R01HG011795 and philanthropic donations to the TCH NRI. We thank the family for their participation.
CONFLICT OF INTEREST STATEMENT
The authors disclose no competing financial conflict of interest.
ETHICS STATEMENT
The participant in this study provided informed consent under the Community Texome project.
Open Research
DATA AVAILABILITY STATEMENT
The data from the case discussed in this report is available on ClinVar under accession number SCV002577663.1.




