In silico validation revealed the role of SCN5A mutations and their genotype–phenotype correlations in Brugada syndrome
Hung Manh Pham and Duy Phuong Nguyen contributed equally to this work.
[Correction added on June 25, 2024 after first online publication. The author names ‘Duy Phuong Nguyen’ and ‘Phong Hai Phan’ has been corrected to ‘Duy Phuong Dang’ and ‘Dinh Phong Phan’ respectively in this version.]
Abstract
Background
Brugada syndrome (BrS) is a rare genetic disease that causes sudden cardiac death (SCD) and arrhythmia. SCN5A pathogenic variants (about 30% of diagnosed patients) are responsible for BrS.
Aims
Lack of knowledge regarding molecular characteristics and the correlation between genotype and phenotype interfere with the risk stratification and finding the optimal treatment in Vietnam. Therefore, we identified SCN5A variants and evaluated the genotype–phenotype correlation of BrS on 117 Vietnamese probands.
Materials and Methods
The clinical characteristics and blood samples of BrS patients were collected. To determine SCN5A variants, Sanger sequencing was conducted, and subsequently, these variants were analyzed by bioinformatic tools.
Results
In this cohort, the overall rate of detected variants in SCN5A was 25.6%, which could include both pathogenic and benign variants. In genetic testing, 21 SCN5A variants were identified, including eight novels and 15 published variants. Multiple bioinformatic tools were used to predict variant effect with c.551A>G, c.1890+14G>A, c.3338C>T, c.3578G>A, and c.5484C>T as benign, while other variants were predicted as disease-causing. The family history of SCD (risk ratio [RR] = 4.324, 95% CI: 2.290–8.269, p < 0.001), syncope (RR = 3.147, 95% CI: 1.668–5.982, p = 0.0004), and ventricular tachycardia/ventricular fibrillation (RR = 3.406, 95% CI: 1.722–5.400, p = 0.0035) presented a significantly higher risk in the SCN5A (+) group, consisting of individuals carrying any variant in the SCN5A gene, compared to SCN5A (−) individuals.
Conclusion
The results contribute to clarifying the impact of SCN5A variants on these phenotypes. Further follow-up studies need to be carried out to understand the functional effects of these SCN5A variants on the severity of BrS.
1 INTRODUCTION
Brugada syndrome (BrS) (OMIM: 600163) is an inherited disorder of cardiac electrical conduction, characterized by a specific ST-segment elevation in the right precordial leads (V1-V3) on the electrocardiogram (ECG), strongly associated with the high risk of ventricular arrhythmias and sudden death (Antzelevitch, 2006). Clinical presentation of BrS also involves sudden infant death syndrome and sudden unexpected nocturnal death syndrome. About 4% of all sudden death cases and at least 20% of sudden death cases with typical heart structures are associated with BrS (Kusano, 2013). The overall prevalence of BrS is globally estimated at 0.05% and found to be of different incidences in different regions. The highest prevalence of BrS is verified in Southeast Asia (3.7 per 1000 population), followed by the Middle East and South Asia, with both rates of 1.8 per 1000 population (Vutthikraivit et al., 2018). Males show a higher prevalence and more severe clinical manifestations than females (Benito et al., 2008). Mutations in SCN5A gene are associated with BrS, accounting for approximately 30% of BrS cases (Brugada et al., 2018). Variety of genes have been identified to be related to BrS, but SCN5A gene is the most dominant when occupied by >5% of positive genotyped patients (Ackerman et al., 2011). According to The European Society of Cardiology guidelines, SCN5A genetic testing is recommended in cases of Class I: the family with carriers of SCN5A mutation in BrS, Class IIa: the patients suspected with BrS based on clinical features, family history, and electrocardiographic (ECG) testing, Class IIb: the patients with ECG type 2 and type 3 of BrS (Priori et al., 2015). The genetic testing in affected family members plays a pivotal role in treatment classification which comprises at-risk (+) and precaution (−) groups.
SCN5A gene is located on chromosome 3p21 and has 28 exons that encode for the cardiac voltage-gated sodium channel (Nav1.5). Nav1.5 contains four transmembrane domains (DI-DIV) connected by inter-domain linkers. Each domain includes six transmembrane segments (S1-S6) joined by intra-extracellular loops (Rook et al., 2012). Segments S1-S4 in each domain are essential to forming a voltage-sensing domain (VSD) that is activated to generate sodium current (Chen-Izu et al., 2015). Nav1.5 induces the depolarization of atrial and ventricular action potentials, thus creating the up-stroke of the cardiac action potential (Jiang et al., 2020). To date, more than 900 SCN5A variants are associated with cardiac disorders and scattered throughout different gene regions. The variants induce the diversity of BrS phenotypes as well as causing loss-of-function or gain-of-function of sodium channel via altered voltage- and time-dependent activation/inactivation and decelerated recovery from inactivation (Savio-Galimberti et al., 2018). Therefore, identifying SCN5A variants and their functional assessment are vital in optimizing and personalizing patient treatment.
In Vietnam, data regarding the molecular characteristics of SCN5A variation are still lacking. Very few studies have determined the association between the genotype and phenotype of SCN5A variants in Vietnamese patients. Recently, computation tools have become widely used to assess the consequence of variants potentially causing disease (Thusberg et al., 2011). Thus, we conducted this study to screen for SCN5A variants and evaluated the genotype–phenotype correlation of BrS on 117 probands. The results will support risk stratification of BrS for treatment and provide detailed information to clinicians for genetic counseling.
2 MATERIALS AND METHODS
2.1 Patients collection
A total of 117 Brugada patients were included from the Vietnam National Heart Institute from January 2017 to April 2022. The patients were diagnosed with BrS according to the diagnostic criteria of the European Heart Rhythm Society. All individuals participated in the current study and signed informed consent.
Clinical characteristics of patients included age, sex, family history of sudden cardiac death (SCD) (<45 years old), clinical treatments, comorbidities, ECG measurements, sodium channel blocker challenge test, and electrophysiological study. This study was approved by the Ethics Committee of Hanoi Medical University (Hanoi Medical University Institutional Review Board, Hanoi, Vietnam, reference number: 48/HĐĐĐĐĐHYHN).
2.2 Molecular genetic analysis
The genomic DNA of each participant was isolated from peripheral blood using a QIAamp Blood Kit (Qiagen) according to the manufacturer's instructions. Then, DNA concentration was measured using NanoDrop 2000 spectrophotometer (Thermo Scientific) and stored at −20°C until use.
The SCN5A coding regions were amplified by PCR using the designed primers at flanking intronic sequences described Park et al. (2012). Twenty microliters of PCR mix contains GoTaq Hot Start Master Mix 2X, 5 pmol primer, DNA template, and distilled water. PCR cycle was carried out by initial denaturation at 95°C for 5 min, followed by 37 cycles of denaturation at 95°C for 30 s, annealing at Tm of primers for 30 s, and extension at 72°C for 30 s, and final extension at 72°C for 5 min. PCR products were observed on 1.5% agarose gel by electrophoresis. Next, PCR amplicons were purified and direct sequenced on ABI 3500 genetic analyzer system (Applied Biosystems). Sequencing data were assembled to reference sequences on NCBI (NG_008934 and NM_198056) to determine the SCN5A variants by CLC Main Workbench 6.0 software.
To determine the large genomic alteration, including deletions and insertions, multiplex ligation-dependent probe amplification (MLPA) was carried out by using the SALSA MLPA probe mix P108-B2 SCN5A (MRC Holland). Results were analyzed with Coffalyser software (MRC Holland).
2.3 Predicting function of SCN5A variants
We analyzed the results on eight computational tools, including six separated predictors: Mutation Assessor (http://mutationassessor.org/r3/), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), Mutation Taster (https://www.mutationtaster.org/), PROVEAN (http://provean.jcvi.org/index.php), FATHMM (http://fathmm.biocompute.org.uk/), SIFT (https://sift.bii.a-star.edu.sg/index.html), and two meta-scores: BayesDel addAF and MetaSVM (Adzhubei et al., 2010; Choi & Chan, 2015; Feng, 2017; Kim et al., 2017; Reva et al., 2011; Schwarz et al., 2014; Shihab et al., 2013; Sim et al., 2012). These algorithms were evaluated based on the structure and function of the targeted protein and evolutionary conservation in its sequence.
2.4 Statistical analyses
A quantitative variable is presented as mean ± standard deviation. Fisher's exact test was calculated to determine the significance of categorical variables. A risk ratio (RR) was applied to determine the correlation between phenotype and genotype of SCN5A variants. A two-tailed p < 0.05 was considered statistically significant. All tests were computed by GraphPad Prism 8 software package (San Diego, USA).
3 RESULTS
3.1 Baseline clinical manifestations
The clinical manifestations and electrocardiographic variables of the 117 participants in this study are summarized in Table 1. Of those participants, males were the majority with 97.4% (114/117) compared to females with 2.6% (3/117). Genetic testing of SCN5A gene categorized the participants into two groups: SCN5A genotype positive and SCN5A genotype negative, accounting for 25.6% and 74.4%, respectively. For the purposes of this analysis, patients with any detected variants in SCN5A were included in the genotype-positive group, including benign variants as well as pathogenic variants. The average age was 47.5 ± 12.4 years, ranging from 23 to 79 years. However, no significant difference was observed in age (p = 0.056), whereas there was a different significance related to family history with sudden death in both groups (p < 0.0001).
| Total cases (n. %) | SCN5A mutation | |||
|---|---|---|---|---|
| Positive (n. %) | Negative (n. %) | p-Value* | ||
| n | 117 | 30 (25.6%) | 87 (74.4%) | |
| Age | 47.5 ± 12.4 | 43.6 ± 10.6 | 48.3 ± 9.8 | 0.056 |
| Family history of sudden cardiac death (<45 years old) | 37 | 20 (66.67) | 17 (19.54) | <0.0001* |
| Clinical manifestations | ||||
| Syncope | 45 (38.46) | 19 (66.33) | 26 (29.89) | 0.002* |
| Nocturnal agonal respiration | 11 (9.4) | 4 (13.33) | 7 (8.05) | 0.4694 |
| Ventricular tachycardia and ventricular fibrillation | 8 (6.83) | 6 (20.00) | 2 (2.30) | 0.0035* |
| Cardiac arrest survivors | 7 (5.98) | 4 (13.33) | 3 (3.45) | 0.0704 |
| Clinical treatments | ||||
| None | 31 (26.5) | 9 (30.00) | 22 (25.28) | 0.6284 |
| Pacemaker implantation | 86 (73.5) | 21 (70.00) | 65 (74.71) | 0.6362 |
| Comorbidities | ||||
| Other arrhythmias | 7 (5.98) | 1 (3.33) | 6 (6.90) | 0.6759 |
| Atrial fibrillation | 4 (3.41) | 1 (3.33) | 3 (3.45) | 1.000 |
| Other cardiovascular diseases | 23 (19.65) | 8 (26.67) | 15 (17.24) | 0.2915 |
| Other internal diseases | 43 (36.75) | 14 (46.67) | 29 (33.33) | 0.1218 |
| Type Brugada in ECG | ||||
| Type 1 | 83 (70.95) | 23 (76.67) | 60 (68.96) | 0.0806 |
| Type 2 | 20 (17.09) | 3 (10.00) | 17 (19.54) | 0.2754 |
| Type 3 | 14 (11.96) | 4 (13.33) | 10 (11.49) | 0.7527 |
| Testing | ||||
| Flecainide testing (+) | 9 (7.69) | 7 (23.33) | 2 (2.30) | 0.001* |
| Electrophysiology study testing (+) | 54 (46.15) | 10 (33.33) | 45 (51.72) | 0.0934 |
- * Statistical significance (p < 0.05).
Of all collected patient features, SCN5A (+) was identified with different rates in syncope, nocturnal agonal respiration, ventricular tachycardia/ventricular fibrillation (VT/VF), and cardiac arrest survivors by 66.33%, 13.33%, 20%, and 13.33%, respectively, compared to 29.89%, 8.05%, 2.3%, and 3.45% of SCN5A (˗). The prevalence of syncope and the incidence of VT/VF presented the statistical difference between SCN5A (+) and SCN5A (˗) groups with p = 0.002 and 0.0035, respectively. Neither clinical treatment methods nor comorbidities and types of BrS in ECG did show any different significance in both groups (p > 0.05). In addition, SCN5A (+) group had a higher rate of flecainide testing than the SCN5A (−) group (23.33% vs. 2.30%; p = 0.001). By contrast, a higher electrophysiology study testing was recorded in SCN5A (−) group than SCN5A (+) group, with the rate of 51.72% and 33.33% (p > 0.05), respectively.
3.2 Association analysis between phenotype and genotype of SCN5A mutation
To assess the risk of SCN5A genotype to BrS phenotype, we evaluated the RR of the clinical characteristics associated with SCN5A mutation. The results are shown in Table 2. The increased risk of 4.324 with a family history of sudden death (<45 years old) in SCN5A (+) probands (95% CI: 2.290–8.269; p < 0.0001) compared to SCN5A (−) probands. The relative risk for syncope, VT/VF, and flecainide testing were also evaluated at 3.147 (95% CI: 1.668–5.982; p = 0.0004), 3.406 (95% CI: 1.722–5.4; p = 0.0035), and 3.652 (95% CI: 1.957–5.737; p = 0.001), respectively, according to SCN5A-positive and -negative groups.
| Relative risk ratio | 95% confidence interval | p-Value* | |
|---|---|---|---|
| Family history of sudden death (<45 years old) | 4.324 | 2.290–8.269 | <0.0001* |
| Syncope | 3.147 | 1.668–5.982 | 0.0004* |
| Ventricular tachycardia and ventricular fibrillation | 3.406 | 1.722–5.400 | 0.0035* |
| Flecainide testing (+) | 3.652 | 1.957–5.737 | 0.001* |
- * Statistical significance (p < 0.05).
3.3 Novel SCN5A variants
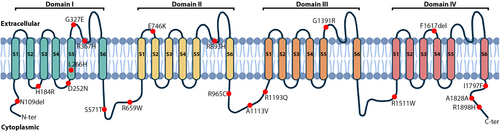
In our study, eight of 21 SCN5A variants were found as novel variants and were not previously recorded in the databases (ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/; LOVD: https://www.lovd.nl/; dbSNP: https://www.ncbi.nlm.nih.gov/snp/). All SCN5A variants are displayed in Table 2 and Figure 1. The location on the protein sequence was followed by the UniprotKB (Q14524), as described: the N-terminus (aa 1–131), domain I (DI S1-S6, aa 132–410), inter-domain linker I-II (aa 411–717), domain II (DII S1-S6, aa 718–938), inter-domain linker II-III (IDL II-III, aa 939–1206), domain III (DIII S1-S6, aa 1207–1466), inter-domain linker III-IV (IDL III-IV, aa 1467–1529), domain IV (DII S1-S6, aa 1530–1771), and the C-terminus (aa 1772–2016). The novel variants were located mainly in the DI, accounting for 15.15%, compared to 6.06% in both N-terminal and C-terminal regions. Among SCN5A variants, a frameshift variant was determined in two patients, such as c.325_327delAAC (N109del) in exon 3. Also, genomic duplication was observed in two patients who showed a duplicate of exon 2 and 3 by MLPA. Five missense variants were identified along 28 exons of SCN5A: c.551A>G (Exon 5), c.754G>A (Exon 7), c.797T>A (Exon 7), c.980G>A (Exon 8), c.5389A>T (Exon 28), produced the amino acid change at His184Arg, Asp252Asn, Leu266His, Gly327Glu, and Ile1797Phe, respectively. A synonymous variant c.5484C>T was found in a single patient at nucleotide position 5484 with no effect on the wild-type 1828 Alanine amino acid residue (Ala1828Ala).
3.4 Known SCN5A variants
Aside from novel variants, 13 known SCN5A variants were recorded in this study. Of these, 11 missense variants have been verified in the dbSNP, containing c.1100G>A (R367H) in exon 9 (rs28937318), c.1712G>C (S571T) in exon 12 (rs199473126), c.1975C>T (R659W) in exon 13 (rs730880205), c.2236G>A (E746K) in exon 14 (rs199473582), c.2678G>A (R893H) in exon 16 (rs199473172), c.2893C>T (R965C) in exon 17 (rs199473180), c.3338C>T (A1113V) in exon 18 (rs199473194), c.3578G>A (R1193Q) in exon 20 (rs41261344), c.4171G>A (G1391R) in exon 24 (rs780405533), c.4534C>T (R1511W) in exon 26 (rs137854602), and c.5693G>A (R1898H) in exon 28 (rs370694515). Two patients carried c.1890+14G>A (rs145427253) at the splice site region of intron 12 and a TCT deletion at the position 4850 to 5852 (c.4850_4852delTCT) (rs749697698), which caused frameshift on protein sequence (I1617del) was found in exon 27.
3.5 In silico analysis of SCN5A variants
To determine the impact of SCN5A variants in BrS, multiple bioinformatics tools were utilized; if the results of ≥4 tools were in agreement on pathogenicity, a variant was considered damaging. The results are revealed in Table 3. Of a total of 21 variants, 15 were predicted as disease-causing by at least four algorithms. Significantly, the variants c.797T>A, c.980G>A, c.1100G>A, c.2678G>A, c.2893C>T, and c.4534C>T were indicated as potentially pathogenic variants by all algorithms. Conversely, five variants were identified as benign, including c.551A>G, c.1890+14G>A, c.3338C>T, c.3578G>A, and c.5484C>T.
| Nucleotide | Protein | Location | Variant type | dbSNP ID | Cases | References |
|---|---|---|---|---|---|---|
| Ex2-Ex3 Dup | Exon 2–3 | Frameshift | - | 2 | In this study | |
| c.325_327delAAC | N109del | Exon 3, N-terminal | Frameshift | - | 2 | In this study |
| c.551A>G | H184R | Exon 5, DI/S2-S3 | Missense | - | 2 | In this study |
| c.754G>A | D252N | Exon 7, DI/S4-S5 | Missense | - | 1 | In this study |
| c.797T>A | L266H | Exon 7, DI/S5 | Missense | - | 1 | In this study |
| c.980G>A | G327E | Exon 8, DI/S5-S6 | Missense | - | 1 | In this study |
| c.1100G>A | R367H | Exon 9, DI/S5-S6 | Missense | rs28937318 | 1 | Kapplinger et al. (2010) |
| c.1890+14G>A | Intron 12 | Splice site | rs145427253 | 2 | Duzkale et al. (2013) | |
| c.1712G>C | S571T | Exon 12, DI-DII | Missense | rs199473126 | 1 | Kapplinger et al. (2009) |
| c.1975C>T | R659W | Exon 13, DI-DII | Missense | rs730880205 | 4 | Shen et al. (2022) |
| c.2236G>A | E746K | Exon 14, DII/S1-S2 | Missense | rs199473582 | 1 | Kapplinger et al. (2010) |
| c.2678G>A | R893H | Exon 16, DII/S5-S6 | Missense | rs199473172 | 2 | Yamagata et al. (2017) |
| c.2893C>T | R965C | Exon 17, DII-DIII | Missense | rs199473180 | 4 | Chimparlee et al. (2021) |
| c.3338C>T | A1113V | Exon 18, DII-DIII | Missense | rs199473194 | 1 | Kapplinger et al. (2010) |
| c.3578G>A | R1193Q | Exon 20, DII-DIII | Missense | rs41261344 | 1 | Abe et al. (2018) |
| c.4171G>A | G1391R | Exon 24, DIII/S5-S6 | Missense | rs780405533 | 1 | Szperl et al. (2018) |
| c.4534C>T | R1511W | Exon 26, DIII-DIV | Missense | rs137854602 | 1 | Meregalli et al. (2009) |
| c.4850_4852delTCT | F1617del | Exon 27, DIV/S3-S4 | Frameshift | rs749697698 | 2 | Chen et al. (2005) |
| c.5389A>T | I1797F | Exon 28, C-terminal | Missense | - | 1 | In this study |
| c.5484C>T | A1828A | Exon 28, C-terminal | Synonymous | - | 1 | In this study |
| c.5693G>A | R1898H | Exon 28, C-terminal | Missense | rs370694515 | 1 | te Riele et al. (2017) |
4 DISCUSSION
BrS is a rare inherited cardiac disorder and is generally related to sudden death during sleeping in South East Asia. Most BrS patients are diagnosed without clarifying clinical manifestations, leading to the challenge of early diagnosis. SCN5A gene encodes for the cardiac voltage-gated sodium channel (Nav1.5) and is responsible for the incidence of BrS. The SCN5A variants are estimated in up to 30% of the patients. Loss-of-function mutations on SCN5A have been investigated to cause dilated cardiomyopathy and lower expression levels of SCN5A or produce defective Nav1.5 proteins (Wilde & Amin, 2018). Therefore, genetic screening of SCN5A is necessary for the clinical diagnosis of BrS patients, identification of non-genetic carriers, and selection of promising strategies to prevent genetic carriers in the offspring.
BrS is well known to be more distributed and severe in males than in females. The vast frequency of affected males in this study was marked at 97.43%, in agreement with prevalence in Asia (Milman et al., 2019). Several hypotheses have been postulated to clarify the gender discrepancy. Variable sex receptors containing estrogen, progesterone, and testosterone are discovered in the cardiac myocytes; thus, the alteration of sex hormone concentration can cause fluctuation in the performance and expression of myocardial ion channels (Lizotte et al., 2009).
In our study, the prevalence of SCN5A variants was determined at 25.6%, higher than Taiwanese and Hispanic populations, which accounted for 8% and 16%, respectively (García-Castro et al., 2010; Juang et al., 2015). Syncope and SCD are common clinical characteristics in BrS-related SCN5A variants. We found that syncope and a risk family history of SCD were presented in 38.46% and 31.62% of our BrS patients, respectively. The syncope symptom of those who carried SCN5A variants (66.33%) has a significantly higher risk than those of non-carriers (RR = 3.147, [95% CI: 1.668–5.982], p = 0.004). A similar rate was obtained in a meta-analysis, which was evaluated at 35.9% in the occurrence of syncope related to SCN5A (+) (RR = 1.12, [95% CI: 0.87–1.45], p = 0.37) (Raharjo et al., 2018). Moreover, our data contrast with the result of Yamagata et al. (2017), who did not observe a statistical difference in the family history of SCD (p = 0.5); however, a significantly higher rate of cardiac events was recorded between SCN5A (+) and SCN5A (−) groups (HR = 1.1, 95% CI: 1.1–3.8, p = 0.02) (Yamagata et al., 2017). In addition, BrS could be related to the major arrhythmic events (MAEs), comprising VT and VF (RR = 3.406, [95% CI: 1.722–5.4], p = 0.0035). Sommariva et al. (2013) indicated that the patients harboring SCN5A variants had an increased risk of MAEs than none of those (p = 0.024) (Sommariva et al., 2013). Nishii et al. (2010) determined the existence of SCN5A variants related to early and frequent recurrence of VF in symptomatic BrS patients, not in the first onset of VF (21.7% vs. 20.0%, p = 0.373) (Nishii et al., 2010). The variable correlation of BrS clinical phenotype may be produced by inter-individual genetic differences, regulating excitation-contraction physiology and affecting arrhythmia susceptibility among distinct ethnicities (Splawski et al., 2002; Suh & Vijg, 2005).
In the current study, 21 different SCN5A variants were successfully identified. Interestingly, one genomic duplication (Ex2-Ex3 dup) (2/117) was detected in our patients. To date, very few publications have shown the large rearrangement in SCN5A. Eastaugh et al. (2011) first described a major deletion in SCN5A. The c.999-424_1338 + 81del causes loss of exon 9 and 10 of SCN5A, resulting in haploinsufficiency (Eastaugh et al., 2011). The deletions of SCN5A were expressed in the other regions such as promoter, exons, and even the whole SCN5A gene (Hertz et al., 2015; Jenewein et al., 2017; Trujillo-Quintero et al., 2019). It is a fact that genomic instability always leads to the complete loss of function of the affected gene; therefore, these variants are more likely to induce the BrS phenotype. Several studies have been performed to identify the genomic imbalance in SCN5A, but the frequency rearrangement was 0% (García-Molina et al., 2013; Koopmann et al., 2007). These findings suggest that MLPA technique should be included when conducting genetic screening for BrS, even though the rearrangement rate is infrequent.
The novel variant c.325_327delAAC (N109del) was located on the N-terminal of Nav1.5 protein. N-terminal is a conserved region and plays a crucial role in regulating the cardiac sodium channel. The alteration of N-terminal can cause the increase or decrease of function on the channel, involving a quality control mechanism (Clatot et al., 2012). Following this mechanism, cells carried mutants use endoplasmic reticulum-associated degradation (ERAD) as a protective role to degrade proteins that fail to acquire their native conformation by the ubiquitin–proteasome system (Brodsky & Scott, 2007). The majority of our novel missense variants were discovered scattered from S2 to S6 segments of DI, consisting of c.551A>G (H184R), c.754G>A (D252N), c.797T>A (L266H), and c.980G>A (G327E). Most of these variants were predicted as pathogenic variants, except c.551A>G (H184R) as benign (Figure 1; Table 4). The S1-S4 components of each domain are considered as the voltage sensor domain (VSD), which plays a central function in controlling the activity of the channel, while S5-S6 segments assemble to form the pore domain and are responsible for the ion selectivity of Nav1.5 (Chen-Izu et al., 2015). Thus, minor structure modifications can initiate the dysregulation functioning of the channel. Additionally, two novel variants, c.5389A>T (I1797F) and c.5484C>T (A1828A), were found on the C-terminal of Nav1.5 and predicted as disease-causing and benign, respectively. The substitutions in the C-terminal domain have been explored to modulate channel inactivation (Glaaser et al., 2006). Although the synonymous c.5484C>T (A1828A) variant did not change the amino acid sequence, it may disrupt mRNA transcription, translation, and splicing (Goymer, 2007; Stergachis et al., 2013).
| Nucleotide change | Variant | In silico analysis tools | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation assessor | Poly-phen2 | Mutation taster | PROVEAN | FATHMM | SIFT | BayesDel addAF | MetaSVM | Total | ||
| Ex2-Ex3 Dup | - | - | - | - | - | - | - | - | - | |
| c.325_327del | N109del | - | - | DC | DC | - | - | - | - | DC |
| c.551A>G | H184R | BN | BN | BN | BN | DC | BN | DC | DC | BN |
| c.754G>A | D252N | BN | DC | DC | DC | DC | BN | DC | DC | DC |
| c.797T>A | L266H | DC | DC | DC | DC | DC | DC | DC | DC | DC |
| c.980G>A | G327E | DC | DC | DC | DC | DC | DC | DC | DC | DC |
| c.1100G>A | R367H | DC | DC | DC | DC | DC | DC | DC | DC | DC |
| c.1890+14G>A | - | - | BN | - | - | - | - | - | BN | |
| c.1712G>C | S571T | BN | DC | DC | BN | DC | BN | DC | DC | DC |
| c.1975C>T | R659W | BN | DC | DC | DC | DC | DC | DC | DC | DC |
| c.2236G>A | E746K | BN | BN | DC | BN | DC | BN | DC | DC | DC |
| c.2678G>A | R893H | DC | DC | DC | DC | DC | DC | DC | DC | DC |
| c.2893C>T | R965C | DC | DC | DC | DC | DC | DC | DC | DC | DC |
| c.3338C>T | A1113V | BN | BN | BN | BN | DC | BN | BN | BN | BN |
| c.3578G>A | R1193Q | BN | BN | BN | BN | DC | BN | BN | BN | BN |
| c.4171G>A | G1391R | BN | DC | BN | DC | DC | DC | DC | DC | DC |
| c.4534C>T | R1511W | DC | DC | DC | DC | DC | DC | DC | DC | DC |
| c.4850_4852del | F1617del | - | DC | DC | DC | - | - | - | - | DC |
| c.5389A>T | I1797F | BN | DC | DC | DC | DC | DC | DC | DC | DC |
| c.5484C>T | A1828A | BN | - | DC | BN | - | BN | - | - | BN |
| c.5693G>A | R1898H | BN | DC | DC | DC | DC | BN | DC | DC | DC |
- Abbreviations: BN, benign; DC, disease-causing; -, not available.
Together with novel variants, 13 known variants also were identified. The c.1100G>A (R367H, rs28937318) variant has been evaluated to cause loss-of-function in Nav1.5 channel and is also associated with atrial standstill and early repolarization in idiopathic ventricular fibrillation (Kapplinger et al., 2010; Takehara et al., 2004; Watanabe et al., 2011). The c.1890+14G>A variant was found in the intronic region and was predicted benign on the ClinVar database (rs145427253) (Duzkale et al., 2013). Other variants were determined to occur at 28.57% (6/21) on the internal domain linker, such as DI-II (c.1712G>C (S571T), c.1975C>T (R659W)), DII-III (c.2893C>T (R965C), c.3338C>T (A1113V), c.3578G>A (R1193Q)), and DIII-IV (c.4534C>T (R1511W)) (Abe et al., 2018; Chimparlee et al., 2021; Kapplinger et al., 2009, 2010; Meregalli et al., 2009; Shen et al., 2022). Of these variants, c.3338C>T (A1113V) and c.3578G>A (R1193Q) were predicted as benign, and damage was evaluated in the other. The mutation rate on the linker domains on Nav1.5 was relatively high, accounting for 35% in the study of Kapplinger et al. (2009) Several studies have reported the affection of SCN5A variants on internal domain linkers. They indicated that these linkers play a valuable function in pathogenesis (Smits et al., 2005; Wehrens et al., 2003; Yamamura et al., 2010). Variant c.2236G>A (E746K) (DII/ S1-S2) was a hot spot for BrS and related to the reduced activation process on Nav1.5 channel (Huang et al., 2017; Zaytseva et al., 2022). An alteration c.2678G>A caused the change of amino acids from Arg to His at position 893, also detected on DII. This variant induced loss-of-function, showing a significant decrease in the current density peak than the wild-type through patch clamp (Ishikawa et al., 2021). The c.4171G>A (G1391R) variant was first reported by Szperl et al. (2018) in the Polish population. Genotype and phenotype analysis showed that the carriers with G1391R variant presented a higher risk of cardiac events in females than in males (Szperl et al., 2018). The minor deletion c.4850_4852delTCT (F1617del) variant was found to be associated with LQT3 syndrome, which induced the change of fast inactivation by decreasing gating charges (Chen et al., 2005). According to te Riele et al. (2017), the missense variant c.5693G>A (R1898H) led to a decrease at 36% in peak sodium current (p = 0.002), also in the abundance of Nav1.5 (p = 0.005) on fluorescence microscopy and N-Cadherin (p = 0.026) clusters at the intercalated disk (te Riele et al., 2017).
Our study's genotype–phenotype correlation and the functional prediction of all variants were only approached by statistical analysis and bioinformatic tools. Further, follow-up studies should be performed to provide insight into the role of SCN5A variants in the severity of BrS and promptly specify the most appropriate treatment for each patient.
5 CONCLUSION
Our study initially evaluated the genotype–phenotype correlation of SCN5A variants with patients of BrS in Vietnam. The presence of SCN5A variants shows a significantly higher risk in the family history of sudden death, syncope, and arrhythmia. Aside from published SCN5A variants, seven additional variants were predicted to cause functional alteration in cardiac voltage-gated sodium channel (Nav1.5). Follow-up studies should be conducted to clarify the impact of SCN5A variants on the clinical characteristics of BrS.
AUTHOR CONTRIBUTIONS
Van-Khanh Tran, Hung Manh Pham, Thinh Huy Tran, Thanh Van Ta, and The-Hung Bui: Conceived and designed the study and analysis. Thi Phuong Le, Dinh Phong Phan, Hoai An Trinh, Thanh Van Ta, and Ha Minh Nguyen: Contributed to data collection and carried out the experiments, performed analysis, and finalized the results. Hung Manh Pham, Duy Phuong Dang, Thanh Dat Ta, and Van-Khanh Tran: Contributed to the drafting of the manuscript. All authors have read and approved the final version for publication.
ACKNOWLEDGMENTS
We sincerely thank the patients and their parents for giving us consent and allowing us to publish the data.
FUNDING INFORMATION
This study was supported by the Ministry of Health of Vietnam, grant number 2723/QĐ-BYT.
CONFLICT OF INTEREST STATEMENT
The authors declare that there is no conflict of interest regarding the publication of this article.
ETHICS STATEMENT
This study was approved by the Ethics Committee of Hanoi Medical University (Hanoi Medical University Institutional Review Board, Hanoi, Vietnam, reference number: 48/HĐĐĐĐĐHYHN).
CONSENT
Written informed consent was obtained from the patient for publication.
Open Research
DATA AVAILABILITY STATEMENT
The data are available from the corresponding author upon request.




