A deleterious frameshift insertion mutation in the ZNF142 gene leads to intellectual developmental disorder with impaired speech in three affected siblings: Clinical features and literature review
Abstract
Background
ZNF142 gene is a protein-coding gene encoding Zinc Finger Protein 142. ZNF proteins are a vast group of cellular effectors with a wide range of functions such as signal transduction, transcriptional regulation, meiotic recombination, DNA repair, development, and cell migration. Mutations in the ZNF142 gene are related to neurodevelopmental disorder with impaired speech and hyperkinetic movements (NEDISHM). This study on a family with three affected siblings identified a pathogenic frameshift insertion variant. In addition, we conducted a review of the literature on previously reported ZNF142 gene variants and their clinical manifestations.
Materials and Methods
Three affected siblings with severe intellectual developmental disabilities and speech impairments, their parents, and other sibs in the family were included. The patients were studied by the whole exome sequencing. Sanger sequencing, co-segregation analysis, and in silico analysis were carried out to verify candidate variant. The identified variant was interpreted based on the ACMG guideline.
Results
We identified a frameshift insertion variant in the ZNF142 gene, NM_001379659.1: c.3755dup (NP_001366588.1:p.Arg1253ThrfsTer15), that was related to the clinical features of three patients. The identified variant was found to be pathogenic.
Conclusion
The current study findings expand the existing knowledge of the variant on the ZNF142 gene implicated in the neurodevelopmental disorder, intellectual disability, and impaired speech and it presents a detailed clinical feature associated with related conditions. The data have implications for genetic diagnosis and counseling in families with the same disorders.
1 INTRODUCTION
The ZNF142 gene (OMIM: 604083; HGNC ID:12927) is a protein-coding gene that encodes Zinc Finger Protein 142. The encoding protein belongs to the zinc-finger protein (ZNF) superfamily. The ZNF superfamily is one of the largest groups of mammalian transcription factors. ZNF proteins contain the Cys2His2 (C2H2) motif coordinating a Zn2+ ion and are known as zinc fingers. They include a large number of cellular effectors with a variety of functions. Several biological functions, including signal transduction, transcriptional control, meiotic recombination, DNA repair, development, and cell migration, are mediated by proteins having zinc-finger domains (Schwarz et al., 2019). Therefore, the ZNF proteins are one of the major transcription factor families in humans, and pathogenic variants in these proteins were reported to be associated with neurodevelopmental disorders (Al-Naama et al., 2020; Tommerup & Vissing, 1995).
ZNF142 is a member of the ZNF family encoded by the ZNF142 gene. The gene is located on 2q35. It consists of 11 exons and encodes 1887 amino acids (Stelzer et al., 2016). The ZNF142 protein includes 31 C2H2-type zinc fingers domains which belong to the Kruppel C2H2-type zinc-finger protein family. The possible role that could be suggested for ZNF142 is the transcriptional regulation (O'Leary et al., 2016). Pathogenic variants in the ZNF142 gene are reported to be related to 10 phenotypes including global developmental delay, ataxia, dystonia, tremor, dolichocephaly, torticollis, delayed speech, and language development, bilateral tonic–clonic seizure, chorea, and hyperkinetic movements (Köhler et al., 2021).
At first, Khan et al. (2019) reported that the ZNF142 gene defects lead to a neurological condition named Neurodevelopmental Disorder with Impaired Speech and Hyperkinetic Movements (NEDISHM; MIM: 618425) (Khan et al., 2019). To our knowledge, our work is the fifth study on the ZNF142 gene related to intellectual disability, and neurodevelopmental disorder with impaired Speech. However, few studies are reported on the clinical spectrum and pathogenic variants of this gene.
In this study, we focused on a family with three affected siblings with severe intellectual developmental disabilities and speech impairments who were found to have a pathogenic frameshift insertion variant. We also thoroughly reviewed the literature on previously reported ZNF142 gene variants and their clinical manifestations.
2 METHODS AND SUBJECTS
2.1 Family recruitment and DNA extraction
The Ethics Committee of the Isfahan University of Medical Sciences, Isfahan, Iran, authorized this research (Ethics code: IR.MUI.MED.REC.1400.042). Three individuals with severe intellectual disability (ID) belonging to a consanguineous family were identified in the Iranian province of Sistan and Balouchestan. Medical history was obtained via genetic counseling, and the pedigree was drawn by the “Progeny” software (Progeny Software, LLC). The peripheral blood was taken after getting informed written consent from the legal guardian. DNA extraction was done using the DNSol Miniprep Kit provided by ROJETECHNOLOGIES company, Tehran, Iran.
2.2 Whole-exome sequencing and variant prioritization
Exome capturing was performed using xGen Exome Research Panel v2 kit (Integrated DNA Technologies, Coralville, Iowa, USA), and whole exome sequencing (WES) was performed using NovaSeq 6000 (Illumina, San Diego, CA, USA) in 3Billion Inc. (Seoul, South Korea). The quality of FASTQ Illumina Novaseq 6000 output files was assessed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Subsequently, the base and sequence adapters with low base quality were removed using Trimmomatic (Bolger et al., 2014). Pre-processed FASTQ files were aligned to the reference sequence by BWA-MEM (v.0.7.17) (Li & Durbin, 2009). Aligned BAM files were sorted and extracted using the statistical metric by samtools (v.1.9) (Li et al., 2009). Duplication was marked by Picard (v.2.20.8) (http://broadinstitute.github.io/picard/). Single nucleotide variants and indel variants were called by HaplotypeCaller of GATK (v.3.8) (McKenna et al., 2010). In total, 9,745,010,591 bases of the sequence were generated and uniquely aligned to the Genome Reference Consortium Human Build 37 (GRCh37) and Revised Cambridge Reference Sequence (rCRS) of the mitochondrial genome, generating 139.03 mean depth-of-coverage within the 34,366,188 bases of the captured region, which is approximately 99.3% of the RefSeq protein coding region. Approximately 98.80% of the targeted bases were covered to a depth of ≥20× (Gene or exon level depth-of-coverage information is available upon request). In total, 67,572 single nucleotide variants (SNV) and 10,888 small insertions and deletions (indel) were identified (The entire variant list without annotation or clinical interpretation is available upon request). The EVIDENCE software (Seo et al., 2020), a software tool developed in-house, has been used to prioritize variants and interpreted based on the guideline recommended by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) (Richards et al., 2015) in the context of the patient phenotypes. Through genetic counseling, pertinent family history and past clinical test findings were supplied. At the time of variant interpretation, only clinically important variations related to the patient's principal clinical indications were examined.
2.3 Sanger sequencing and co-segregation analysis
The candidate variant was confirmed using Sanger sequencing, and co-segregation analysis was performed on affected and unaffected members of family. Specific primers for the variant were designed using the Primer3 online tool (Primer3web, version 4.1.0) and validated by online tools such as Primer-BLAST (Ye et al., 2012), MFEprimer3.1 (Wang et al., 2019) and SNPCheck (gene tools, SNPCheck V3). The used primer sequences are listed in Table 1.
| Primer name | Sequence (5′ – > 3′) | Product size | Genomic coordinate (GRCh37) |
|---|---|---|---|
| Forward | 5′- CGCTTCTGGTGGAGCCTTA −3′ | 406 bp | chr2:219507771–219508177 |
| Reverse | 5′- TTGAGCAGGGCAAGTTTCAC -3’ |
2.4 In silico analysis
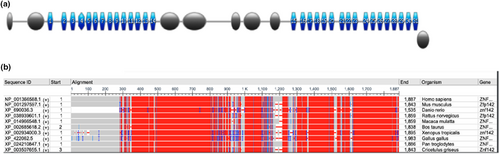
We used the Genome Aggregation Database (gnomAD v2.1.1) and Iranome (http://www.iranome.ir/) for population allele frequency analysis. The potential pathogenicity of the variants was assessed using the NMDEscPredictor server (https://nmdprediction.shinyapps.io/nmdescpredictor/). The NCBI Constraint-based Multiple Alignment Tool (COBALT) server (version 1.22.0) was used for the analysis of conservation (https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?CMD=Web). ACMG classification was performed by using the Varsome online tool (https://varsome.com/). The schematic view of the ZNF142 protein was provided by the Prosite protein database (https://prosite.expasy.org/).
3 RESULTS
3.1 Clinical findings
The proband is a 23-year-old girl with severe ID and speech disorder (Figure 1a-Individual IV-3). She was born at term after an uncomplicated pregnancy. She is the third child of a consanguineous family with Balouch ethnicity from Iran. Although birth parameters and neonatal adaption were normal, she had delayed in milestones. She lifts their head by 4 months old, sit with some support by 8 months, and started walking at 24 months. Her speech development was impaired; she spoke her first two- to three–3-word sentences at the age of five. She could not speak clearly and only used some unclear words at the time of this study. As described, she had a neurodevelopmental delay during infancy. She suffers from a learning disability that worsens with her age and is considered as a severe ID. She also suffered from enuresis since childhood. She did not encounter mobility challenges, seizures, and other medical problems. She shows no dysmorphic features. She had a normal number of chromosomes based on cytogenetics and normal aCGH test results. She has two younger siblings with the same conditions. Her younger brother is 17 years old (Figure 1a-Individual IV-4) and suffers from severe ID and speech impairment, he had a neurodevelopmental delay, learning disability, and enuresis. In general, he is similar in terms of signs and symptoms to the proband. They also have a younger 7-year-old brother (Figure 1a-Individual IV-5) with moderate ID and speech impairments. He is similar in conditions to his two elder affected siblings but did not show developmental delay. His phenotype is milder than his two siblings. There is a family history of the same conditions in their father's cousins that did not want to participate in the study (Figure 1a-Individual III-1,2,3).
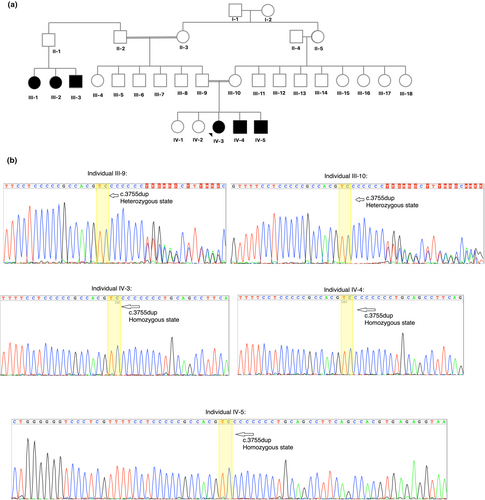
3.2 Molecular and in silico findings
Sanger sequencing and segregation analysis validated the NM_001379659.1 (ZNF142):c.3755dup homozygous frameshift insertion variant in the ZNF142 gene (Gene Bank ID: GC02M218637) that was identified using WES (Figure 1b). The variant was found to be homozygous in the three afflicted individuals, and both parents were carriers for the variant. The variant is observed at an extremely low frequency in the gnomAD dataset (total allele frequency: <0.001%) and not found in the Iranome dataset (Table 2). Some in silico tools also demonstrated the variant negative influence on gene function, such as the NMDEscPredictor server which predicted to result in a loss or disruption of normal protein function through nonsense-mediated decay (NMD) and protein truncation (Figure 2) and COBALT server showed the locus of the variant and downstream regions are highly conserved in multiple organisms (Figure 3) that their deletion could affect protein functions. ACMG classification of the variant was pathogenic based on PVS1 (null variant), PM2 (variant not found in gnomAD genomes), and PP1 (co-segregation with the disease in multiple affected members of the family) lines of evidence (Table 2).
| Gene | Genomic position | cDNA position | Protein change | gnomAD frequency | Iranome frequency | Pathogenicity line of evidence based on ACMG |
|---|---|---|---|---|---|---|
| ZNF142 | 2-219,508,083-T-TC (GRCh37) | NM_001379659.1: c.3755dup | NP_001366588.1:p.Arg1253ThrfsTer15 | <0.001% | Not found | PVS1 (null variant), PM2 (variant not found in gnomAD genomes), and PP1 (co-segregation with the disease in multiple affected members of the family) |

4 DISCUSSION
The ZNF142 gene defect was first described by Khan et al. (2019) related to a specific neurodevelopmental condition. They discovered homozygous or compound heterozygous mutations in the ZNF142 gene lead to “Neurodevelopmental Disorder with Impaired Speech and Hyperkinetic Movements” (NEDISHM; MIM: 618425). It is an autosomal recessive disorder characterized by global developmental delay apparent in infancy. Khan et al. studied seven patients with NEDISHM and autosomal recessive inheritance belonging to the four unrelated families (Khan et al., 2019). The majority of their studied patients have modest delays in walking and speaking, cognitive impairment, speech deficits, and motor impairment, in addition to hyperkinetic movement abnormalities such as dystonia, tremors, ataxias, and chorea. They found some individuals may develop seizures that gradually diminish (Table 3) (Khan et al., 2019). In 2021 Kameyama et al. described a Malaysian family with two affected members suffering from global developmental delay, epilepsy, dysmorphism, and typical facial features (Table 3) (Kameyama et al., 2022).
| Family | Patient | Variant description | Variant type | Inheritance | Protein change | Protein domain | Clinical findings | Method | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | A | NM_001105537.2: c.817_818del/c.1292delG | Frameshift/Frameshift | Compound heterozygote | p.(Lys273Glufs*32)/p.(Cys431Leufs*11) | C2H2-type 3/C2H2-type 9 | Mild ID, Speech impairment, Developmental delay, Ataxia, Tremor, Dystonia, Dolichocephaly, Torticollis | Whole exome sequencing | Khan et al. (2019) |
| B | Mild ID, Speech impairment, Developmental delay, Ataxia, Tremor, Dystonia, Dolichocephaly, Torticollis, Seizure, Behavior problems, Facial dyskinesia, Movement disorder | Whole exome sequencing | |||||||
| 2 | A | NM_001105537.2: c.3175C > T | Nonsense | Homozygote | p.(Arg1059*) | Region | Moderate ID, Speech impairment, Developmental delay, Ataxia, Seizure, Hypotonia, Signal intensities in brain MRI, Dystonia, Choreatic movements, excessive beta activity in EEG, Torticollis, Excessive crying | Whole exome sequencing | Khan et al. (2019) |
| 3 | A | NM_001105537.2: c.3698G > T/c.4498C > T | Missense/Missense | Compound heterozygote | p.(Cys1233Phe)/p.(Arg1500Trp) | C2H2-type 18/C2H2-type 26 | Mild ID, Speech impairment, Developmental delay, Brain MRI: Mild thinning of the posterior aspect of the body of the corpus callosum with an associated mild reduction in white matter volume, brisk reflexes, Increased tone and reflexes in lower limbs | Whole exome sequencing | Khan et al. (2019) |
| 4 | A | NM_001105537.2: c.4183_4185delinsAT | Nonsense | Homozygote | p.(Leu1395*) | C2H2-type 23 | Severe ID, Speech impairment, Seizure, Dolichocephaly | Whole exome sequencing | Khan et al. (2019) |
| B | Severe ID, Speech impairment, Developmental delay, Ataxia, Tremor, Seizure | Whole exome sequencing | |||||||
| C | Severe ID, Speech impairment, Developmental delay, Tremor, Dolichocephaly, Seizure | Whole exome sequencing | |||||||
| 5 | A | NM_001105537.4: c.1252C > T/c.1274-2A > G | Nonsense/Nonsense | Compound heterozygote | p.(Arg418*)/p.(Glu426*) | C2H2-type 8/- | Severe ID, Speech impairment, Developmental delay, Seizure, Hypotonia, Bilateral mesial temporal sclerosis in brain MRI, Autistic features, Dolichocephaly, Prominent eyes, Micrognathia, Flat nasal bridge, Thick lips, Accessory nipple, Arachnodactyly | Whole exome sequencing | Kameyama et al. (2022) |
| B | Moderate ID, Speech impairment, Developmental delay, Seizure, Delayed myelination in brain MRI, Autistic features, Atonia, Dolichocephaly, Prominent eyes, Micrognathia, Flat nasal bridge, Prominent Forehead, Caf ́e au lait spot | Whole exome sequencing | |||||||
| 6 | A | NM_001105537.2: c.25C > T/c.2288C > G | Nonsense/Missense | Compound heterozygote | p.(Gln9*)/p.(Ser763Cys) | Region/Region | Mild ID, Seizures, Abnormal EEG, Broad nasal bridge, Wide nose, Narrow palpebral fissures, Syndactyly of 2nd and 3rd toe, Amenorrhea | Trio whole genome sequencing | Christensen et al. (2022) |
| 7 | A | NM_001105537.2: c.527del/c.4030C > T | Frameshift/Nonsense | Compound heterozygote | p.(Thr176Ilefs*93)/p.(Arg1344*) | C2H2-type 1/C2H2-type 21 | Neurodevelopmental delay, Speech impairment, Moderate ID, Behavioral problems, Dystonia, Tremor, Ataxia, Hypotonia, Hyperkinetic Movements, Seizures, Thick lips, Faunal ears, Oblic palpebral fissures, Overlapping teeth, Narrow palate, Strabismus, Long and bulging chest, Nipples apart, Scoliosis and kyphosis, Hands and feet laxity, Clinodactyly on feet | Whole exome sequencing |
Christensen et al. (2022) |
| 8 | A | NM_001105537.2: c.1165_1166del/c.1910del | Frameshift/Frameshift | Compound heterozygote | p.(Asp389Serfs*9)/p.(Pro637Leufs*23) | C2H2-type 7/Region | Neurodevelopmental Delay, Speech Impairment, Moderate ID, Autistic behavior, Seizure, MRI: Small degree of cranial asymmetry, Hypotonic facies with open mouth, Pes planus, Joint laxity, Ataxia | Targeted exome sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental Delay, Speech Impairment, Mild ID, Autistic behavior, Seizure, Hypotonic facies with open mouth, Pes planus, Joint laxity | Sanger sequencing | |||||||
| 9 | A | NM_001105537.2: c.1252C > T | Nonsense | Homozygote | p.(Arg418*) | C2H2-type 8 | Neurodevelopmental Delay, Speech Impairment, Severe ID, Autistic behavior, Tremor, Mild hypotonia, Complex movement disorder, Dysarthria, Dysphagia, Salivation, Axial rigor, Brady diadochokinesia, Mild postural instability. Stereotypic hand movements, Seizure, Abnormal EEG, Hypomimia, Macrocephaly (56 cm), Dwarfism, Mild thoracic scoliosis. Pes planus, Male-pattern hypertrichosis (face, legs, genital region) | Sanger sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental Delay, Speech Impairment, Moderate ID, Mildly ataxic gait, Mild hypotonia, Seizure, Joint laxity, Renal involvement | Whole exome sequencing | |||||||
| 10 | A | NM_001105537.2: c.1456C > T | Nonsense | Homozygote | p.(Gln486*) | C2H2-type 11 | Neurodevelopmental Delay, Speech Impairment, Moderate ID, Unexplained laughing, Dysonia, Tremor, Ataxia, MRI: Thin corpus callosum, deep white matter dysmyelination around the occiptial horn of lateral ventricles, mild cortical involution with deep Sylvian fissures and deep interhemispheric fissure (3 years), High forehead, Wide palpebral fissures, Braod nose, Long philtrum, Thin Upper lip, Everted lower lip, Pointed chin, Low set ears | Whole exome sequencing |
Christensen et al. (2022) |
| 11 | A | NM_001105537.2: c.1906C > T/c.3735del | Nonsense/Nonsense | Compound heterozygote | p.(Arg636*)/p.(Leu1245Phefs*4) | Region/C2H2-type 18 | Neurodevelopmental Delay, Speech Impairment, Autism, Mild Hypotonia, Cerebral visual impairment, Delayed myelinization parieto-occipital, Thick eyebrows, Mild synophrys, Broad nasal bridge, Epicantal folds, High nasal bridge, Long and broad philtrum, Hair on upper lip, Ears are normally implanted, folded but posteriorly rotated, Proximal thumbs, Hockey stick palmar crease unilaterally, Long and broad first rays of the feet; inspection of the mouth, nek, upper body is normal, Primary amenorrhea | Trio exome sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental Delay, Speech impairment, ADHD, Cerebral visual impairment, Small cyst with surrounding gliosis at the tip of the anterior horn of the left lateral ventricle with contiguous prominent vascular structure in the caput Nuclei caudati, crus anterior of capsule interna right down to the nucleus lentiformis, Thick eyebrows, Mild synophrys, Broad nasal bridge, Hypertelorism, Epicantal folds, Mild bilateral ptosis, High nasal bridge, Long and broad philtrum, Ears are normally implanted, folded but posteriorly rotated | Direct analysis of the familial variant | |||||||
| 12 | A | NM_001105537.2: c.1906C > T/c.3885C > A | Nonsense/Missense | Compound Heterozygote | p.(Arg636*)/p.(Phe1295Leu) | Region/C2H2-type 20 | Speech impairment, Episodic ataxia, Neonatal hypertonia, Seizure, High-arched palatum, Systolic cardiac souffle | Whole exome sequencing |
Christensen et al. (2022) |
| 13 | A | NM_001105537.2: c.2650del | Frameshift | Homozygote | p.(His884Thrfs*3) | – | Speech impairment, Moderate ID, Dystonia, Tremor, High forehead, Thick eyebrows, Epicanthus, Thick lips, Posteriorly rotated ears | Whole exome sequencing, Sanger sequencing |
Christensen et al. (2022) |
| 14 | A | NM_001105537.2: c.2851del/c.3167dup | Frameshift/Frameshift | Compound heterozygote | p.(Glu951Lysfs*66)/p.(Gly1057Argfs*11) | Region/region | Neurodevelopmental Delay, Speech Impairment, Moderate ID, Behavior problem, Sleep disorder, Hypotonia, Seizure, Mild hypertelorism, Overfolded helices, Narrow palate, Overlapping toe, Clinodactyly | Whole exome sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental Delay, Speech Impairment, Mild ID, Mild peripheral hypertonia, Axial hypotonia, Seizure, Bilateral epicanthus, Wide nasal bridge, Anteverted nares, Pronounced philtrum, Overfolded helices, Left single palmar crease | Whole exome sequencing | |||||||
| 15 | A | NM_001105537.2: c.3155dup | Frameshift | Homozygote | p.(Arg1053Thrfs*15) | Region | Speech impairment, Moderate ID | Whole exome sequencing |
Christensen et al. (2022) |
| B | Speech impairment, Severe ID, Narrow forehead, Short philtrum, Thick lips | Sanger sequencing | |||||||
| C | Speech impairment, Moderate ID, Narrow forehead, Long face, Long nose, Thick lips | Sanger sequencing | |||||||
| 16 | A | NM_001105537.2: c.3175C > T | Nonsense | Homozygote | p.(Arg1059*) | Region | Neurodevelopmental Delay, Speech Impairment, Global developmental delay, Broad-based walking, Seizure, Facial hypotonia, Thick lips, Dolicocephaly, Retrognathia, Curled ears | Whole exome sequencing |
Christensen et al. (2022) |
| 17 | A | NM_001105537.2: c.3346del | Frameshift | Homozygote | p.(Glu1116Asnfs*4) | – | Speech impairment, Moderate ID, Social disorder, Behavior problems, Seizure, Macrocephaly, Long face, Epicanthus, Low-set ears, Joint laxity, Arachnodactyly, Long and narrow feet, Long toes, Unilateral cryptorchidism | Trio exome sequencing |
Christensen et al. (2022) |
| 18 | A | NM_001105537.2: c.3514C > T | Nonsense | Homozygote | p.(Gln1172*) | C2H2-type 16 | Neurodevelopmental delay, Speech impairment, Mild ID, Aggressive Behavior, Seizure, Abnormal EEG, Kyphosis | Sanger sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental delay, Speech impairment, Moderate ID, Pes planus | Whole genome sequencing | |||||||
| C | Neurodevelopmental delay, Speech impairment, Moderate ID, Tremor, Seizure, Pes planus | Whole genome sequencing | |||||||
| 19 | A | NM_001105537.2: c.4261C > T | Nonsense | Homozygote | p.(Gln1421*) | – | Neurodevelopmental delay, Speech impairment, Mild ID, Anxiety, Aggressive behavior, Ataxia, Hypotonia, Seizure, Abnormal EEG, High forehead, Thick lips, Scoliosis, kyphosis, Strabismus, Hypermetropia | Whole exome sequencing |
Christensen et al. (2022) |
| 20 | A | NM_001105537.2: c.4436del | Frameshift | Homozygote | p.(Pro1479Leufs*45) | – | Neurodevelopmental delay, Speech impairment, Mild ID, Dystonia, Hypotonia, Seizure, Long thin face, Broad nasal root and tip, Narrow palate | Whole exome sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental delay, Speech impairment, Mild ID, Some immature behaviors | Whole exome sequencing | |||||||
| 21 | A | NM_001105537.2: c.4440C > G | Nonsense | Homozygote | p.(Tyr1480*) | C2H2-type 26 | Neurodevelopmental delay, Speech impairment, Moderate ID, Dystonia, Hypotonia, Seizure, Abnormal EEG, Narrow forehead, Thick eyebrows, Narrow palpebral fissures, Hanging upper eyelids, Broad nasal bridge, Prominent nose with hypoplastic nares, Very short philtrum, Everted lower lip, Prominent incisors, Hyperlordosis and scoliosis, Slender and long arms, Short stature, Myopia, Pubertas tarda | Whole exome sequencing |
Christensen et al. (2022) |
| B | Neurodevelopmental delay, Speech impairment, Moderate ID, Hypotonia, Seizure, Long face, Medial flaired eyebrows, Thick eyebrows, Short philtrum, Thick lips, Prominent incisors, Uvula bifida (lower part), Dental crowding, Low set ears | Whole exome sequencing | |||||||
| 22 | A | NM_001105537: c.25C > T/c.1741C > T | Nonsense/Missense | Compound heterozygote | p.Gln9*/p.Arg581Cys | Region/C2H2-type 14 | ID, Speech impairment, Developmental delay, Seizure, Hypotonia | Whole exome sequencing | Kamal et al. (2022) |
| 23 | A | NM_001379659.1: c.3755dup (NM_001105537.2: c.3155dup) | Frameshift | Homozygote | p.Arg1253ThrfsTer15 | Region | Severe ID, Speech disorder, Developmental Delay | Whole exome sequencing | Present Study |
| B | Sanger sequencing | ||||||||
| C | Sanger sequencing |
Christensen et al. (2022) described 16 families containing 26 patients with NEDISHM and autosomal recessive inheritance (Table 3). The majority of the studied patients show intellectual disability, language impairments, and delay in developmental milestones, and some show seizures, hypotonia, behavioral problems, movement disorders, dystonia, tremor, and ataxia (Christensen et al., 2022). Kamal et al. (2022) reported an Iranian family with mutations in the ZNF142 gene in an affected child with intellectual disability, developmental delay, hypotonia, and seizures (Table 3) (Kamal et al., 2022). Approximately 36 people in four studies have been diagnosed with NEDISHM to yet, and the present investigation identified another family with three afflicted siblings who have the ZNF142 gene mutation and suffer from neurodevelopmental delay, severe intellectual disability, and speech dysfunction.
The NM_001379659.1 (ZNF142):c.3755dup homozygous variant was detected in all three sibs of the current study, whose parents were carrier heterozygous. The variant is located in exon 9 of 11 and in a region between two C2H2 domains. The identified variant is located in a region consisting of seven C nucleotide repeats, and therefore replication slippage mechanism can lead to the insertion of additional C nucleotides in that position (Viguera et al., 2001). The variation alters the Arginine residue at residue 1253 of the protein at the protein level. After 15 amino acids, a premature stop codon is formed, resulting in a shortened protein lacking highly conserved domains (Figure 3b). Therefore, the last two exonic regions of the gene encoding the critical C2H2 domains are lost. Hence, multiple pathogenic variants were reported downstream of this variant. In addition to the missed functional domains, the transcript could be exposed to the nonsense-mediated decay process, which shortens the half-life of the transcript (Holbrook et al., 2004). Moreover, the identified deleterious variant, NM_001379659.1 (ZNF142):c.3755dup, was recently reported from Iran on the same ethnicity group related to the same condition (Christensen et al., 2022), which might implicate it as a variant of the founder effect. In general, along with the current study, about 29 variants in this gene have been reported, which are related to neurodevelopmental delay conditions (Table 3). Based on HGMD non-professional, 16 out of the 29 variants are base substitutions and 14 are INDELs. Thus, the frequency of point substitution and small insertion/deletion variants are almost equal. Therefore, loss-of-function (LOF) is a known mechanism of disease (gene has 15 reported pathogenic LOF variants). Besides, compound heterozygous variants should be considered in the ZNF142 gene variants analysis (Table 3) (Schaaf et al., 2012).
According to previous studies, pathogenic variants in this gene resulted in a spectrum of clinical presentations including intellectual disability, speech impairment, developmental delay, hyperkinetic movements, ataxia, tremor, dystonia, dolichocephaly, torticollis, seizure, deficits in social and executive functions, Facial dyskinesia, movement disorder, hypotonia, atonia, some facial features. Along with other studies, the patients in current study show intellectual disability, neurodevelopmental delay, delayed milestones, and speech impairments. Nevertheless, despite the chosen term for the linked disorders, hyperkinetic movements were not seen in the current investigation or the majority of prior studies. Table 3 presents comparing of reported variants and their detailed clinical manifestations with the current study. It is concluded that different types of mutations, either homozygous or compounds heterozygous on C2H2 and regions could lead to various phenotypes. However, more studies are needed to conclude a genotype–phenotype correlation.
5 CONCLUSION
In conclusion, this study revealed a recurrent deleterious variant in the ZNF142 gene (NM_001379659.1 (ZNF142):c.3755dup), identified by WES, and described clinical findings in the family. The results broaden the mutational and clinical spectrum of the ZNF142 gene.
AUTHOR CONTRIBUTIONS
Atefeh Mir: Patient identification and provision of clinical data, exome data analysis, and mutation screening, performing experiments and segregation analysis in families by Sanger sequencing, and writing the first draft of the manuscript. Yongjun Song: Performing whole exome sequencing and data analysis. Hane Lee: Performing whole exome sequencing and data analysis. Mostafa Montazer-Zohouri: Recruitment of patients and family members and sampling, performing experiments. Marziyeh Reisi: Recruitment of patients and family members and sampling, performing experiments. Mohammad Amin Tabatabaiefar: Design and supervision of the research, review and editing the first draft of the manuscript.
ACKNOWLEDGMENTS
We sincerely thank the family who participates in this study. We are sincerely grateful to 3Billion Inc., Seoul, South Korea, for performing the whole exome sequencing. The Isfahan University of Medical Sciences also supported the study, Isfahan, Iran (Grant No. 3991102). This is a part of A. M.'s Ph.D. thesis.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
ETHICS STATEMENT
The research was performed in accordance with the approval of the ethics board of the Medical University of Isfahan (Ethics code: IR.MUI.MED.REC.1400.042).
Open Research
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article and the raw data that support the findings of this study are available from the corresponding author upon reasonable request. In addition, the identified variant during the current study has been submitted in the ClinVar repository with accession number; VCV001705330.1.




