(Epi)genetic profiling of extraembryonic and postnatal tissues from female monozygotic twins discordant for Beckwith–Wiedemann syndrome
Laura Fontana and Maria F. Bedeschi have contributed equally to this work.
Abstract
Background
Beckwith–Wiedemann syndrome (BWS) is an overgrowth disorder caused by defects at the 11p15.5 imprinted region. Many cases of female monozygotic (MZ) twins discordant for BWS have been reported, but no definitive conclusions have been drawn regarding the link between epigenetic defects, twinning process, and gender. Here, we report a comprehensive characterization and follow-up of female MZ twins discordant for BWS.
Methods
Methylation pattern at 11p15.5 and multilocus methylation disturbance (MLID) profiling were performed by pyrosequencing and MassARRAY in placental/umbilical cord samples and postnatal tissues. Whole-exome sequencing was carried out to identify MLID causative mutations. X-chromosome inactivation (XCI) was determined by HUMARA test.
Results
Both twins share KCNQ1OT1:TSS-DMR loss of methylation (LOM) and MLID in blood and the epigenetic defect remained stable in the healthy twin over time. KCNQ1OT1:TSS-DMRLOM was nonhomogeneously distributed in placental samples and the twins showed the same severely skewed XCI pattern. No MLID-causative mutations were identified.
Conclusion
This is the first report on BWS-discordant twins with methylation analyses extended to extraembryonic tissues. The results suggest that caution is required when attempting prenatal diagnosis in similar cases. Although the causative mechanism underlying LOM remains undiscovered, the XCI pattern and mosaic LOM suggest that both twinning and LOM/MLID occurred after XCI commitment.
1 INTRODUCTION
Beckwith–Wiedemann syndrome (BWS, OMIM #130650) is an imprinting disorder primarily characterized by pre- and post-natal overgrowth and an increased risk of developing embryonal tumors (Weksberg, Shuman, & Smith, 2005). Additional clinical features of BWS include the following: neonatal hypoglycemia/hyperinsulinism, macroglossia, facial nevus simplex, ear creases/pits, abdominal wall defects/omphalocele, visceromegaly, and hemihyperplasia (lateralized overgrowth). BWS is a pan-ethnic condition, with an estimated prevalence of 1:10,500 live births (Mussa et al., 2013) and an equal incidence in males and females, except for monozygotic twins.
The molecular defects underlying the syndrome are heterogeneous, involve the imprinted 11p15 chromosomal region (Li, Squire, & Weksberg, 1998), and are often present in a mosaic pattern. The variable proportions of mutated cells among tissues is possibly associated with different levels of phenotypic severity. The 11p15.5 chromosomal band harbors two imprinting control regions, H19/IGF2:IG-DMR and KCNQ1OT1:TSS-DMR, which regulate the expression of the IGF2/H19 and CDKN1C/KCNQ1OT1 imprinted domains, respectively. Epigenetic alterations are the leading mechanism causing BWS, with loss of methylation (LOM) at KCNQ1OT1:TSS-DMRaccounting for approximately 60% of cases and gain of methylation at H19/IGF2:IG-DMR in around 10% of patients (Ibrahim et al., 2014). In addition, chromosome anomalies (1%–2%) (Sait et al., 1994; Slavotinek, Gaunt, & Donnai, 1997), segmental paternal uniparental disomy (15%–25%), and maternally derived CDKN1C mutations (5% and 30%–50% of sporadic and inherited cases, respectively), have been also described (Bliek et al., 2001; DeBaun et al., 2002; Weksberg et al., 2001). Together, these defects account for approximately 85% of diagnosed or suspected cases of BWS. Diagnosis of BWS is primarily based on clinical evaluation. The cardinal signs of BWS are considered key features for clinical diagnosis, according to the criteria proposed by Weksberg (Weksberg, Shuman, & Beckwith, 2010) and recently revised by Brioude (Brioude et al., 2018), whereas suggestive features add to the likelihood of a clinical diagnosis and are indicative for molecular testing, but are less specific.
In addition to (epi)genetic alterations at the BWS locus, approximately 25%–50% of patients with BWS exhibit methylation defects extending to other imprinted loci throughout the genome, referred to as multilocus imprinting disturbance (MLID) (Bens et al., 2016; Bliek, Verde, et al., 2009; Fontana et al., 2018; Maeda et al., 2014). Most BWS patients with MLID carry LOM at KCNQ1OT1:TSS-DMR (Arima et al., 2005; Bens et al., 2016; Bliek, Verde, et al., 2009; Court et al., 2013; Docherty et al., 2014; Hiura et al., 2012; Lim et al., 2009; Poole et al., 2013; Rossignol et al., 2006), while the phenomenon is very rare among cases with gain of methylation at H19/IGF2:IG-DMR (Maeda et al., 2014). Some patients with BWS and MLID show loss/gain of methylation at both maternal and paternal iDMRs (imprinted Differentially Methylated Regions) (Court et al., 2013; Maeda et al., 2014), while others have a hypomethylation syndrome restricted to maternally imprinted genes (Baple et al., 2011; Boonen et al., 2008; Sano et al., 2016). The phenotypic consequences of MLID in patients with BWS are disputed, since most patients do not show peculiar phenotypic features, probably as a consequence of the (epi)dominance of one locus above the others (Azzi et al., 2009), or as a result of mosaicism. The etiology of MLID remains largely unknown, although causative mutations in genes involved in DNA methylation maintenance in the zygote/early embryo have rarely been identified in patients with BWS or their mothers (Docherty et al., 2015; Fontana et al., 2018; Meyer et al., 2009). Identification of genetic defects underlying MLID may help to define the recurrence risk of BWS, as previously described (Begemann et al., 2018), and facilitate patient-tailored follow-up.
Peculiarly, among patients with BWS, the incidence of female monozygotic (MZ) twins is increased over that in the general population (2.5% vs. 0.3%–0.4%) (Bliek, Verde, et al., 2009; Weksberg et al., 2002; Hall, 2003). Importantly, twins with BWS are frequently phenotypically discordant, despite the healthy twin also carrying an 11p15.5 alteration, most oftenKCNQ1OT1:TSS-DMR LOM (Bliek, Alders, et al., 2009; Inoue et al., 2017; Tierling et al., 2011; Weksberg et al., 2002); however, the epigenetic defect is only shared in the blood, whereas, the alteration is widespread in other compartments, for example, buccal cells, in the affected twin. Although it has been claimed that exchange of blood cells through placental vascular connections can explain the presence of affected cells in the healthy twin, Bliek, Alders, et al. (2009) proposed another mechanism in which the methylation failure precedes, and possibly triggers, the twinning process itself, and where migration of blood cell precursors from the yolk sac to both twins could explain the presence of hypomethylated cells in the phenotypically healthy twin (Hall, 1996). In addition, MLID is more common in MZ twins discordant for BWS (Bliek, Alders, et al., 2009; Tierling et al., 2011). Bliek, Alders, et al. (2009) reported that approximately 50% of MZ twins with KCNQ1OT1:TSS-DMRLOM showed extended LOM at other imprinted loci.
Regarding the clinical presentation of MZ twins discordant for BWS, the unaffected twin may have some mild manifestations associated with BWS (Orstavik, Tommerup, Eiklid, & Heleneörstavik, 1995; Olney, Buehler, Waziri, Optiz, & Reynolds, 1988), while in BWS-concordant twin pairs, one twin often appears to be more severely affected than the other (Clayton-Smith, Read, & Donnai, 1992), suggesting a broad range of phenotypic presentation. The variable degree of phenotypic discordance in BWS multiple pregnancies has been suggested to be related to the degree of mosaicism, which, in turn, is associated with the timing of the twinning event and establishment of the epigenetic defect (Cohen et al., 2019). According to the algorithm for clinical management of multiple pregnancies (in which at least one child is affected by BWS) recently proposed by Cohen (Cohen et al., 2019), in monozygotic gestations (monochorionic or dichorionic), the patient requiring medical attention should be clinically and molecularly evaluated, while their twin should be evaluated by abdominal ultrasound and clinically examined by a geneticist. Based on clinical score (≥4) and the presence of cardinal BWS features, tumor surveillance is then recommended.
The frequent occurrence of mosaicism in patients with BWS, and its relationship with the phenotypic expression of the disease in single pregnancies, has led to speculation about whether the timing of twinning is related to the expression of the epigenetic defect in multiple gestations. According to the theory of “diffuse mosaicism” (Cohen et al., 2019), the epigenetic event leading to BWS appears to trigger the twinning process; affected cells diffuse in the embryos during multiple gestations, leading to a mosaic distribution that is likely responsible for the phenotypic discordance between twins.
The increased prevalence of female twins in the BWS population suggests a mechanism that triggers epigenetic post-zygotic events forcing the twinning process, specifically in the presence of an XX chromosome complement. According to the Bestor theory (Bestor, 2003), failure of DNA methylation enzymes, mainly DNMT1o (the oocyte form of DNMT1), to maintain KCNQ1OT1:TSS-DMRmethylation may lead to epigenetic asymmetry and the separation, by the twinning event, of cell populations with different KCNQ1OT1:TSS-DMRmethylation patterns. DNMT1o is also involved in X-chromosome inactivation (XCI), possibly linking nonrandom X-inactivation to BWS discordance (McGraw et al., 2013). Although many cases have been published (Bliek, Alders, et al., 2009; Inoue et al., 2017; Tierling et al., 2011; Weksberg et al., 2002), the origin and dynamics of the process leading to twinning discordance for BWS remain unsolved; in addition, no extensive analysis of embryonic tissues has been performed to shed light on the distribution of the epimutation in the extraembryonic compartment.
Here, we report a molecular characterization of extraembryonic and postnatal tissues from two female MZ twins, discordant for BWS and sharing KCNQ1OT1:TSS-DMRLOM and MLID in the blood. In addition, we performed whole-exome sequencing (WES) analysis to identify possible MLID-causative defects. Both twins underwent extensive clinical evaluation and follow-up.
2 MATERIALS AND METHODS
2.1 Patients
The female MZ, monochorionic diamniotic twins were the second-born children of healthy, unrelated Italian parents. At birth, the mother was 36, the father was 46, and the healthy brother was 5 years old. The pregnancy was spontaneously conceived but complicated by twin-to-twin transfusion syndrome, where the twin affected with BWS (S1) was the recipient, and the phenotypically healthy twin (S2) was the donor. Fetoscopic laser coagulation was conducted at the 25th week of gestation (WOG).
The female twins were delivered at 33 WOG + 5 by Caesarean section, due to premature rupture of the membranes. S1 had an APGAR score of 7/9 and S2 8/9 at 1 and 5 min.
Both twins were clinically evaluated for growth at birth, according to twin growth charts, and clinically scored for BWS, according to the recently revised criteria (Brioude et al., 2018). Additional clinical evaluations were conducted at birth, including cardiac, abdominal and cerebral ultrasound, in addition to brain magnetic resonance imaging (MRI). Further, both sisters were regularly monitored at multidisciplinary follow-up, with pediatric, clinical genetic, psychiatric, and neuropsychiatric evaluations until the age of 3 years.
The research study was approved by the Ethical Committee of the IRCCS Ca’ GrandaOspedale Maggiore Policlinico (No. 526/2015).
2.2 DNA extraction and bisulfite modification
Postnatal DNA samples were obtained from peripheral blood leukocytes (PBLs) and buccal swabs (BS) from S1 and S2 at 6, 12, and 18 months of age. PBL DNA samples were also obtained from the parents.
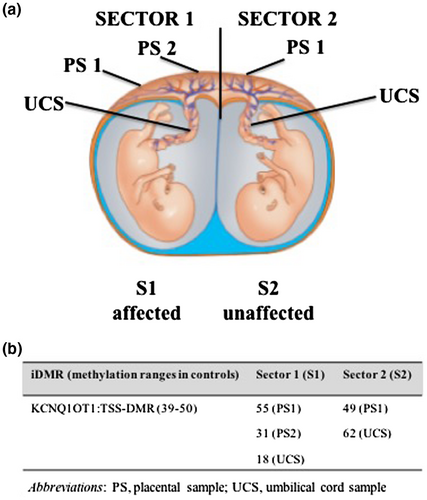
For prenatal retrospective evaluation of extraembryonic tissues, we used paraffin-embedded samples collected at delivery as part of routine pathologist evaluation of complicated pregnancies. Three placental samples, taken from two regions referred to as sector 1 (near to the umbilical cord of S1), and one region from sector 2 (near to the umbilical cord of S2), were investigated, as well as umbilical cord samples from both twins (Figure 1a).
DNA was extracted from fresh and formalin-fixed paraffin-embedded (FFPE) tissue samples using the QIAamp DNA Mini Kit (Qiagen) and the FFPE QIAamp DNA Extraction Kit (Qiagen), following the manufacturer's instructions. S1 and S2genomic DNA samples (500 ng) were subjected to bisulfite-conversion using the EZ DNA Methylation-Gold Kit (Zymo Research).
2.3 Pyrosequencing of the chromosome 11p15 region
Methylation levels at H19/IGF2:IG-DMR and KCNQ1OT1:TSS-DMR in PBLs, BS and FFPE tissues were determined as previously described (Calvello et al., 2013). Quantitative DNA methylation analyses were performed using the Pyro Mark ID instrument (Qiagen) in the PSQ HS 96 System (Qiagen), with the PyroGold SQA reagent kit (Qiagen). The percentage of methylation at each CpG site was automatically calculated using Q-CpG software v1.0.9 (Biotage). Each sample was run in duplicate, and reported methylation values are the mean of at least two independent PCR and pyrosequencing experiments.
Methylation was measured in FFPE tissues (i.e., placental and umbilical cord fragments), since previous studies demonstrate that FFPE treatment does not interfere with DNA methylation patterns (Fontana et al., 2016; Sirchia et al., 2016).
The DNA methylation levels were investigated at H19/IGF2:IG-DMR and KCNQ1OT1:TSS-DMR that have been demonstrated to be germ line DMRs. Given that, DNA methylation is expected to be established in the germ line and maintained through fertilization and development in all somatic lineages (Farhadova, Gomez-Velazquez, & Feil, 2019), and consequently to be very stable and homogeneous among different tissues. For this reason, reported methylation values were expressed as percentages, and the results were interpreted according to methylation ranges previously determined in PBLs, as follows: H19/IGF2:IG-DMR, 40%–52%; KCNQ1OT1:TSS-DMR, 39%–50% (Calvello et al., 2013).
2.4 Copy number variations and methylation status of the chromosome 11p15 region determined by methylation-specific multiplex-ligation probe amplification
An MCR-Holland kit, ME-030 BWS/RSS (MRC Holland), was used following the manufacturer's instructions. The DNA samples were processed in parallel, with and without digestion using the methylation-sensitive HhaI restriction enzyme, to detect both methylation alterations and Copy Number Variations (CNVs) at the chromosome 11p15 genomic region. Data were analyzed using Coffalyser software v. 131211 (MRC Holland).
2.5 MLID assessment by MALDI-TOF mass-spectrometry
The methylation profiles of 12 iDMRs frequently involved in BWS with MLID were investigated using the MassARRAY methylation platform (Agena Bioscience), as recently described by Fontana et al., 2018. MLID analysis was performed on S1 and S2 DNAs from PBLs and BS, after bisulfite modification, as previously described. The Aberrant methylation of each iDMR was defined as an observed methylation value outside two standard deviations from the mean methylation determined in controls, as previously reported (Fontana et al., 2018).
2.6 chromosome inactivation (XCI)
XCI patterns were assessed in PBLs from S1, S2, and their parents, and in BS samples from S1 and S2, by analysis of the methylation status of the androgen receptor gene (HUMARA test) using previously described procedures (Salsano et al., 2012; Sharp, Robinson, & Jacobs, 2000). PBL and BS samples from the twins used for XCI analysis were sampled at the time of molecular diagnosis (6 months after birth). Briefly, PCRs were performed before and after enzyme digestion using the HpaII and HhaI enzymes (Boehringer Ingelheim). Samples were tested in duplicate, with one male sample included in each experiment to verify the enzymatic digestion efficiency. PCR products were separated by capillary electrophoresis, and XCI values were determined using a previously reported formula and definitions (Manoukian et al., 2013). XCI patterns were classified as follows: random, XCI ≤ 75%; moderately preferential, 75% < XCI < 90%; and severely preferential, XCI ≥ 90%.
2.7 WES
The SureSelect Clinical Research Exome V6 Kit (Agilent) was used to perform WES of PBL samples from S1 and her parents. Libraries were run on an Illumina Nextseq 550 (Illumina), sequences were aligned using the BaseSpace BWA Enrichment v2.1 App (Illumina) (through BWA-MEM v0.7.7 on GRCh37/hg19 and Picard v1.79), and variants were called using GATK HaplotypeCaller v1.6. Variants were then annotated using ANNOVAR (April 2018) and filtered according to a custom pipeline (guided by GATK Best Practice), which selectively removed reads with poor quality call scores, low coverage (<10), falling in segmental duplication regions (SuperDups > 0.9), synonymous/unknown variants, and common SNPs (MAF > 1%). Trio analysis was then performed to collect all autosomal dominant/recessive and X-linked recessive variants, as well as de novo variants (File S1).
3 RESULTS
3.1 Clinical data and follow-up
The clinical characteristics of both twins at birth and at 3 years are summarized in Table 1. Their birth weights were in the 75–95th (S1) and 25th (S2) percentiles, while birth lengths were in the 50–75th (S1) and 25th (S2) percentiles, and head circumferences were in the 50–75th (S1) and 25th (S2) percentiles, based on twin growth charts (Kosińska, Sierzputowska-Pieczara, & Gadzinowski, 2018). At birth, the appearance of S1 was characteristic of BWS, with macroglossia and nevus flammeus (score = 3, according to the criteria of Brioude et al., 2018), although there was no macrosomia (birthweight >2 standard deviations above mean), hypoglycemia, or hemihyperplasia/lateralized overgrowth. At 3 years old, S1 was 15 kg (75th centile) and 98 cm tall (90th centile), and her occipital frontal circumference (OFC) was 49 cm (75th centile). In addition, S1 showed facial nevus flammeus and increasing macrosomia. S2 was 11 kg (3rd centile) and 91 cm tall (25th centile) with an OFC of 47.5 cm (25th centile), and absence of any BWS features was confirmed. No lateralized overgrowth was noted in S2 during follow-up, whereas S1 showed lower limb asymmetry (approximately 1.5 cm), which was not observable at birth, but became evident from 3 years of age.
| S1 (affected) | S2 (unaffected) | |||
|---|---|---|---|---|
| Birth | 3 years | Birth | 3 years | |
| Weight, kg (centile) | 2.450 (75–90th) | 15.5 (75th) | 1.895 (25th) | 11 (5th) |
| Length, cm (centile) | 44 (50–75th) | 98 (90th) | 43 (25th) | 91 (25th) |
| OFC, cm (centile) | 32 (50–75th) | 49 (75th) | 30.5 (25th) | 47.5 (25th) |
| Macroglossia | + | + | − | − |
| Exomphalos | − | − | − | − |
| Lateralized overgrowth | − | + | − | − |
| Multifocal and/or bilateral Wilms tumor or nephroblastoma | − | − | − | − |
| Hyperinsulinism (>1 week) | − | − | − | − |
| Adrenal cortex cytomegaly/placental mesenchymal dysplasia/pancreatic adenomatosis | − | − | − | − |
| Facial nevus simplex | + | + | − | − |
| Polyhydramnios and/or placentomegaly | − | − | ||
| Ear creases and/or pits | − | − | − | − |
| Transient hypoglycemia (<1 week) | − | − | − | − |
| Typical BWS tumors | − | − | − | − |
| Nephromegaly and/or hepatomegaly | − | − | − | − |
| Umbilical hernia and/or diastasis recti | − | − | − | − |
| Neurodevelopmental delay | + | + | − | − |
- Abbreviations: BWS, Beckwith–Wiedemann syndrome; OFC, occipital frontal circumference.
S2 did not show any clinical features of BWS. Additional clinical investigations performed after birth showed no major malformations in either twin.
In the context of clinical and instrumental investigations routinely performed for prematurity and for monitoring possible effects of the fetoscopic laser coagulation, brain MRI and neuropsychiatric evaluations were performed in both twins. No defects or malformations were observed on MRI in both twins. Neuropsychiatric evaluation revealed global neurodevelopmental delay in S1, while S2 showed normal psychomotor development, although at the lower limit. Griffiths mental development scale assessment at 25 months revealed GQ values of 67 and 78 in S1 and S2, respectively. In particular, S1 showed delays in all investigated areas (GQ, 76; locomotor quotient, 70; personal and social quotient, 64; language and communication quotient, 50; eye and hand co-ordination quotient, 54; performance quotient, 60). Impairment in praxis, manipulation skills, and hand/eye coordination were also observed. Regarding language and communication, expressive language was more compromised than receptive language; however, macroglossia was not severe enough to cause the language delay. Her communicative intentionality was sometimes poor. Neurological assessment did not reveal any specific neurological deficits or impairment except clumsiness. In conclusion, the observed mild global developmental delay in S1 could be likely caused by prematurity.
S1 underwent neuro-psychomotor and logopedic therapies, particularly for the language delay and sialorrhea, due to macroglossia. S1 also had sleeping problems, including snoring and sleep apnea; polysomnographic evaluation was performed at 10 and 20 months and showed mild to moderate apnea, without impairment of oxyhemoglobin saturation, mainly related to macroglossia. Both twins underwent oncologic surveillance, with abdominal ultrasound and serum alpha fetoprotein level monitoring every 3 months.
3.2 Evaluation of chromosome 11p15 methylation profiles in extraembryonic and postnatal tissues
H19/IGF2:IG-DMRand KCNQ1OT1:TSS-DMR methylation levels were first investigated by pyrosequencing of PBL samples collected from S1 and S2 at 6 months old for diagnostic purposes, when the twins came to our attention for the clinical diagnosis of BWS of S1. In S1, H19/IGF2:IG-DMRmethylation was within the normal range (mean methylation value, 44%), whereas KCNQ1OT1:TSS-DMRwas hypomethylated (mean methylation level, 14%), confirming the clinical diagnosis of BWS in S1, which was related to LOM atKCNQ1OT1:TSS-DMR. KCNQ1OT1:TSS-DMRwas also hypomethylated (mean methylation value, 15%) in PBLs from S2, despite the absence of BWS clinical features (Table 2).
| iDMR (methylation % in controls) | Tissue | Time (months) | S1 (affected) | S2 (unaffected) |
|---|---|---|---|---|
| H19/IGF2:IG (40–52 PBLs) | Blood | 6 | 44 (2.1) | 45 (1.2) |
| KCNQ1OT1:TSS (39–50 PBLs) | Blood | 6 | 14 (1.2) | 15 (0.2) |
| Buccalswab | 25 (2.1) | 45 (3.5) | ||
| Blood | 12 | — | 15 (0.7) | |
| Buccalswab | — | 49 (1.9) | ||
| Blood | 24 | — | 19 (2.7) | |
| Buccalswab | — | 45 (0.9) |
Note
- Reported data are means of independent experiments; standard deviations are reported in brackets.
- Hypomethylated loci are reported in bold and underlined.
- Abbreviations: BS, buccal swabs; PBL, peripheral blood leukocytes.
KCNQ1OT1:TSS-DMRLOM was confirmed in BS samples from S1 (mean methylation value, 26%), whereas methylation was normal in S2 BS specimens (mean methylation value, 45%) (Table 2). There was no evidence of methylation defects at H19/IGF2:IG-DMR in BS from either twin. The KCNQ1OT1:TSS-DMRmethylation profile was also monitored at 12 and 18 months of age in both PBL and BS samples from the non-affected twin (S2); KCNQ1OT1:TSS-DMRvalues remained stable in both PBLs and BS (Table 2).
Methylation-specific multiplex-ligation probe amplification (MS-MLPA) confirmed the pyrosequencing results as follows: S1 showed KCNQ1OT1:TSS-DMRLOM in PBL and BS samples (mean methylation values PBLs = 17% and BS = 32%), while S2 was hypomethylated at KCNQ1OT1:TSS-DMRonly in PBLs (mean methylation values PBLs = 16% and BS = 52%). MS-MLPA also provided no evidence for genomic imbalances/CNVs at the 11p15 locus, suggesting that the KCNQ1OT1:TSS-DMRLOM observed in both twins was due to a primary epigenetic defect.
Pyrosequencing analysis of the placenta showed heterogeneous methylation (Figure 1b). In sector 1 fragments (close to the site of umbilical cord insertion in S1), we found hypomethylation in two of three fragments from two cotyledons, one corresponding to the placental tissue (PS2 = 31%) and the other to the umbilical cord sample (UCS = 18%) (Figure 1b). Placental and umbilical cord samples corresponding to sector 2 (close to the umbilical cord insertion in S2) did not show hypomethylation at KCNQ1OT1:TSS-DMR (PS1 = 49% and UCS = 62%) (Figure 1b). These results show that, in this pregnancy, the placenta had epigenetic mosaicism, with a preponderance of cells affected by KCNQ1OT1:TSS-DMRLOM in the area corresponding to the insertion site of the umbilical cord of the affected twin. These data confirm the epigenetic mosaicism of the placenta and again raise the question as to whether chorionic villi are suitable for use in prenatal diagnosis of BWS (Eggerman et al., 2016).
3.3 Assessment of MLID and underlying genetic defects
Since up to 50% of patients with BWS show MLID, we investigated the presence of epimutations at other iDMRs using a targeted MassARRAY panel including 12 imprinted loci, as reported by Fontana et al. (2018), to analyze PBL and BS samples from both twins at 6 and 24 months. As reported in Table 3, both twins showed MLID, with LOM at multiple maternally iDMRs. Nevertheless, the iDMRs involved were heterogeneous in S1 and S2. S1 showed MLID in PBLs, with LOM at DIRAS-CG1, GNAS1A, PEG10, FAM50B, and in BS at only GNAS2A. S2 had MLID in only PBLs, with LOM at two of the loci affected in S1 (DIRAS-CG1 and FAM50B) and in two loci that were methylated normally in S1 (DLK1 and DIRAS-CG2). These results suggest that MLID can involve different methylation sites in twins, as expected for a stochastic event.
| iDMR (methylation % in controls) | Tissue | S1 | S2 |
|---|---|---|---|
| DIRAS-CG2 (42–72) | Blood | 45 | 38 |
| Buccal swab | 47 | 52 | |
| DIRAS-CG1 (46–66) | Blood | 37 | 33 |
| Buccal swab | 47 | 61 | |
| DLK1 (52–67) | Blood | 58 | 51 |
| Buccal swab | 57 | 59 | |
| GNAS1A (39–51) | Blood | 20 | 45 |
| Buccal swab | 28 | 39 | |
| GNASXL (39–56) | Blood | 45 | 45 |
| Buccal swab | n.a. | n.a. | |
| MEG3 (37–54) | Blood | 44 | 40 |
| Buccal swab | 41 | 40 | |
| MEST (41–66) | Blood | 49 | 44 |
| Buccal swab | 43 | 60 | |
| PEG10 (47–60) | Blood | 46 | 49 |
| Buccal swab | n.a. | n.a. | |
| PLAGL1 (46–58) | Blood | 55 | 54 |
| Buccal swab | n.a. | n.a. | |
| FAM50B (35–47) | Blood | 24 | 26 |
| Buccal swab | n.a. | n.a. |
Note
- Hypomethylated loci are shown in bold and underlined.
- Abbreviations: BS, buccal swabs; n.a., not available; PBLs, peripheral blood leucocytes.
To investigate mutations that may be causative of MLID, we performed WES on samples from S1 (since we confirmed that the twins were MZ by microsatellite genotyping, data not shown) and the parents. We detected a total of 54,751 variants, with a mean Q30 value of 93% and uniform coverage >100×, of which 426 remained after quality control filtering. We focused on analysis of variants in genes with a function in methylation establishment/maintenance; however, we did not find any pathogenic variants potentially correlated with MLID in S1 (File S1). In addition, the mother harbored no variants in maternal-effect genes (Begemann et al., 2018) that could be suspected to lead to MLID in the twins.
3.4 XCI patterns
Since a skewed XCI profile has been hypothesized as being associated with phenotypic discordance in MZ twins with BWS (Orstavik et al., 1995; Weksberg et al., 2002), we investigated XCI patterns in both PBLs and BS collected at 6 months from S1 and S2. In PBLs, both twins had the same XCI pattern, with severe preferential inactivation of the paternally inherited X-chromosome (allele ratios, 99.3%:0.7% for S1 and 99.2%:0.8% for S2). Further, XCI analysis in BS showed similar results (allele ratio: 83%:17% for S1 and 82%:18% for S2). No variants on the X-chromosome possibly associated with the observed skewing of X-inactivation were identified by WES sequencing in S1 (File S1), nor were genomic imbalances affecting the X-chromosome identified by SNParray analysis (data not shown). These data are consistent with previous reports (Weksberg et al., 2002) and indicate that skewing of X-inactivation may be associated with monozygotic twinning, but not with the BWS phenotype.
4 DISCUSSION
The incidence of MZ twins is increased among patients with BWS, relative to the general population (2.5% vs. 0.3%–0.4%) (Bliek, Alders, et al., 2009; Weksberg et al., 2002). It is peculiar that twins are generally phenotypically discordant for a clinical diagnosis of BWS, but share KCNQ1OT1:TSS-DMRLOM in PBLs (Chien, Lee, & Tsai, 1990; Clayton-Smith et al., 1992; Orstavik et al., 1995; Olney et al., 1988), probably as a consequence of sharing of blood circulation, which is a common feature of monozygotic twinning (Weksberg et al., 2002).
We investigated a pair of female twins discordant for BWS by analyzing extraembryonic and postnatal DNA samples collected from different tissues and at follow-up. We first examined the methylation levels at H19/IGF2:IG-DMRand KCNQ1OT1:TSS-DMR, and found LOM at KCNQ1OT1:TSS-DMRin blood from both twins, whereas this locus was only hypomethylated in BS from the affected twin (S1), confirming that hypomethylation can be carried in blood cells without a pathological phenotype, probably as a consequences of placental anastomosis leading to shared blood precursors carrying the epigenetic defects and continuing to differentiate into blood lineages (Bliek, Alders, et al., 2009; Inoue et al., 2017; Tierling et al., 2011; Weksberg et al., 2002). After birth, at 6, 12, and 18 months, we monitored the methylation profiles in blood and buccal smears from the healthy twin (S2), confirming unchanged KCNQ1OT1:TSS-DMR LOM in the blood, without any evident phenotypic effect. Differently from previous reports (Bliek, Alders, et al., 2009), we found that MLID, despite being present in blood samples from both twins, involved different iDMRs in the siblings; moreover, in the affected twin, MLID showed a different pattern in the blood and BS. This evidence suggests that, although no data are available on the distribution of iDMRs involved in MLID at single cell resolution, the mechanisms leading to MLID may affect each cell in a stochastic way, which could result in a nonhomogeneous MLID pattern among different tissues.
As reported in previous studies (Begemann et al., 2018; Sparago et al., 2019), MLID can be associated with maternally inherited (as well as maternal) mutations in genes involved in DNA methylation dynamics; however, in this family we did not identify possible MLID-causative genetic variants by WES of PBL samples from the parents and S1. The tissue distribution of the KCNQ1OT1:TSS-DMRLOM, along with the MLID pattern in both twins, suggests that the mechanism leading to the epigenetic defects acts either after the twinning event (thereby excluding involvement of variants in maternal-effect genes) and/or forces segregation of affected cells to only one blastomere. It could be speculated that the discordant phenotype is related to the asymmetric distribution of epimutated cells. According to the theory of diffuse mosaicism (Cohen et al., 2019), in this pregnancy the twinning event could have occurred immediately after establishment of the epigenetic defect, leading to less time for division and dispersion of affected cells, ultimately resulting in phenotypic discordance. In addition, given the comparable distribution of KCNQ1OT1:TSS-DMRLOM and MLID in the affected tissues, we can speculate that a common causative mechanism underlies the epigenetic defects.
There are two previous reports of twins with BWS and MLID (Bliek, Alders, et al., 2009; Tierling et al., 2011). Bliek, Alders, et al. (2009) only investigated MLID in blood samples from the twins, and identified a comparable hypomethylation in both maternally and paternally iDMRs in a subset of analyzed twins. Unlike Tierling et al., (Tierling et al., 2011) reported no methylation changes at imprinted loci, other than KCNQ1OT1:TSS-DMR in the blood, fibroblasts, saliva, and BS, from a male MZ twin pair discordant for BWS. In contrast, in our twin pair, MLID presented as a maternal hypomethylation syndrome, with a different methylation pattern in each twin and at each analyzed iDMR. In particular, the slight hypomethylation levels retrieved at some iDMRs, such as LOM at DLK1 and PEG10 in S1 and DIRAS-CG2 in S2, relative to other analyzed loci, support the hypothesis that the mechanism underlying MLID acts as a stochastic event affecting different loci in each cell. MLID thus presents as a highly complex form of mosaicism. In the present study, the origin of LOM at iDMRs remains unsolved; however, it was not related to inheritance of novel or already reported variants in known maternal-effect genes that have a possible effect on establishment/maintenance of imprinting in the oocyte (Begemann et al., 2018).
We also retrospectively investigated placental and umbilical cord samples taken from multiple areas corresponding to each twin. This allowed us to determine the distribution of KCNQ1OT1:TSS-DMRLOM in extraembryonic tissues, findings that inform understanding of the distribution of epigenetic defects in twins discordant for BWS. We found that placental KCNQ1OT1:TSS-DMRmethylation levels were heterogeneous within the extraembryonic organ, since only two of three fragments from cotyledons close to the affected twin's umbilical cord insertion were hypomethylated, whereas all samples chosen close to the healthy twin's umbilical cord insertion showed normal KCNQ1OT1:TSS-DMRmethylation. In addition, the umbilical cord sample from S1, but not that from S2, displayed KCNQ1OT1:TSS-DMR LOM. These data suggest that KCNQ1OT1:TSS-DMRLOM is not present in extraembryonic tissues of the phenotypically normal twin, again posing the question of whether KCNQ1OT1:TSS-DMR LOM and MLID triggered the twinning process through an almost completely asymmetric distribution of affected cells. In addition, the heterogeneous distribution of hypomethylation in chorionic villi should be considered if prenatal diagnosis is performed in similar pregnancies. Mosaicism represents a challenge for both clinical and molecular diagnosis, since affected individuals may have heterogeneous clinical presentation and may not be referred for appropriate molecular testing (Mackay & Temple, 2017). For this reason, BWS has been redefined as a spectrum disorder (Beckwith–Wiedemann spectrum or BWSp), with low clinical scores triggering molecular testing and allowing the diagnosis of atypical cases (Brioude et al., 2018). Molecular mosaicism may also hamper accurate diagnosis when molecular testing is performed only on PBLs. For patients with a strong clinical suspicion of BWS, it is mandatory to test other tissues (Azzi et al., 2009). The challenges related to detection of mosaicism already evident in single pregnancies are increased in pregnancies involving twins with suspected BWS; this is because the variable mosaic distribution complicates the prediction of pregnancy outcome, as mosaicism might be confined to the placenta or to the fetus, or it could affect both. Indeed, there can be considerable epigenetic variation within the placenta, suggesting that stochastic and localized effects in the uterine environment may have key roles. It has been hypothesized that this variability may be a function of the number of trophoblast stem cells from which the placental trophoblast derives, with placentae derived from fewer precursors having a greater variance (Katari et al., 2009). For this reason, the irregular distribution of affected cells in the placenta may lead to false-negative results. Another issue in prenatal molecular testing of imprinting disorders using chorionic villus sampling (CVS) is related to the possibility that methylation marks may not be completely set in the placenta at the time of prenatal diagnosis, as discussed by Eggerman et al. (2016), thus making it even more challenging to test for BWS by CVS. We previously confirmed stable methylation levels at the primary imprinting centers, including KCNQ1OT1:TSS-DMR, also in placenta at early stages; therefore, these loci represent reliable targets for prenatal methylation testing of BWS by CVS. This finding is not shared by other investigated loci, thus prenatal investigation of genomic imprinting in CVS should be validated in a locus-specific manner (Paganini et al., 2015).
We also performed molecular analyses of XCI patterns in the twins and detected an almost identical pattern of severe preferential XCI in both children. Indeed, almost 100% of cells showed the same XCI pattern, in which the maternal X was active and the paternal X inactive. Notably, twins were concordant for XCI patterns and discordant for iDMR methylation patterns, indicating that, in this case, establishment of XCI preceded the event leading to LOM at KCNQ1OT1:TSS-DMR and MLID. It is possible that a wave of demethylation occurred after establishment of XCI. This could also explain the complex mosaicism in the iDMR methylation patterns observed in the placenta and between the twins. The presence of a completely skewed XCI pattern in the twins is probably not related to the twinning process itself, since other studies found random patterns of XCI in healthy twins, as well as in those discordant for BWS (Weksberg et al., 2002; Wong et al., 2011). Preferential XCI has been instead associated to some female predominant diseases (Invernizzi, Selmi, Miozzo, Podda, & Invernizzi, 2008). Several hypotheses have been proposed to explain the origin of phenotypic discordance between twins with BWS, which account for the time of the twinning event and the establishment of the imprinting error (Bestor, 2003). In our case, the sharing of the same pattern of skewed X inactivation by the twins suggests that XCI commitment occurred very early in the embryo and remained stable after the twinning process, whereas the presence of nonhomogeneous KCNQ1OT1:TSS-DMRLOM and MLID patterns indicates that the imprinting perturbation was a later event.
In conclusion, this is the first reported case of female twins discordant for BWS in which postnatal tissues were investigated over time and methylation analyses extended to extraembryonic tissues. We showed that the placental distribution of hypomethylated KCNQ1OT1:TSS-DMRcells was heterogeneous, suggesting that caution is required when attempting prenatal diagnosis based on chorionic villi samples in similar cases. Finally, the presence of identical, extremely skewed, XCI patterns in both twins, along with LOM with a mosaic pattern, suggests that both twinning and LOM at imprinted loci occurred after XCI commitment.
ACKNOWLEDGMENTS
We thank Dr Claudia Farè for technical support. This work was supported by the Italian Ministry of Health (Grant RF-2013-02359454) and Fondazione IRCCS Ca' GrandaOspedale Maggiore Policlinico (Grant 5X1000-519 02 and Ricerca Corrente-519 03 to MM). This study has been performed within the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (ERN-ITHACA).
CONFLICT OF INTEREST
None declared.
AUTHORS’ CONTRIBUTION
FL, conceived the experimental plan, coordinated experiments, and drafted the manuscript; BMF, patients’ clinical diagnosis, clinical data management, and manuscript preparation; CGA, PN, GS, PM, and APF clinical data collection and patients’ follow-up, revision of the state-of-art of similar cases; CJ, NGS data analysis; CP, technical support for MLPA and pyrosequencing experiments; SC, WES library preparations and NGS runs; CP, SNParray experiments; SSM, experimental plan of X-chromosome inactivation and supervision of the manuscript; MM, corresponding author and project coordination; ST, supervised the project and contributed to the interpretation of the results.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.