A novel splicing pathogenic variant in COL1A1 causing osteogenesis imperfecta (OI) type I in a Chinese family
Yaxin Han and Dongming Wang are contributed equally as first author.
Abstract
Background
Osteogenesis imperfecta (OI), a rare autosomal inheritable disorder characterized by bone fragility and skeletal deformity, is caused by pathogenic variants in genes impairing the synthesis and processing of extracellular matrix protein collagen type I. With the use of next-generation sequencing and panels approaches, an increasing number of OI patients can be confirmed and new pathogenic variants can be discovered. This study sought to identify pathogenic gene variants in a Chinese family with OI I.
Methods
Whole-exome sequencing was used to identify pathogenic variants in the proband, which is confirmed by Sanger sequencing and cosegregation analysis; MES, HSF, and Spliceman were used to analyze this splicing variant;qRT-PCR was performed to identify the mRNA expression level of COL1A1 in patient peripheral blood samples; Minigene splicing assay was performed to mimic the splicing process of COL1A1 variants in vitro; Analysis of evolutionary conservation of amino acid residues and structure prediction of the mutant protein.
Results
A novel splicing pathogenic variant (c.3814+1G>T) was identified in this OI family by using whole-exome sequencing, Sanger sequencing, and cosegregation analysis. Sequencing of RT-PCR products from the COL1A1 minigene variant reveals a 132-nucleotide (nt) insertion exists at the junction between exons 48 and exon 49 of the COL1A1 cDNA. Splicing assay indicates that the mutated minigene produces an alternatively spliced transcript which may cause a frameshift resulting in early termination of protein expression. The molecular analysis suggested that the altered amino acid is located at the C-terminus of type I procollagen.
Conclusion
Our study reveals the pathogenesis of a novel COL1A1 splicing pathogenic variant c.3814+1G>T in a Chinese family with OI I.
1 INTRODUCTION
Osteogenesis imperfecta (OI), also known as brittle bone disease, is a rare genetic disorder characterized by increased bone fragility, skeletal deformities, and growth deficiency (Forlino & Marini, 2016). Extraskeletal manifestations are highly variable including the blue sclera, joint hyperlaxity, dentinogenesis imperfecta (DI), and hearing loss (Pollitt et al., 2006). Based on the severity of bone fragility, clinical presentation, and radiographic features, OI was classified into four types (types I–IV; OI I, OMIM#166200; OI II, OMIM #166210; OI III, OMIM #259420; OI IV, OMIM #166220) by Silence in the late 1970s (Sillence, Senn, & Danks, 1979).
According to the Sillence's classification system, all OI types from I to IV were caused by abnormal type I collagen or quantitative changes of the collagen. Type I collagen, the main protein building block in bone and other connective tissue, is a triple helix with nonhelical terminal peptides-telopeptides. The triple helix is folded by two pro-α1(Ⅰ)-collagen chains and one pro-α2(Ⅰ)-collagen chain which is encoded by COL1A1 (OMIM #120150) and COL1A2 (OMIM #120160), respectively (Lu, Zhang, Wang, Ren, & Han, 2019). The structure of triple helix is Gly-X-Y repeats with glycine in every third position, and proline and hydroxyproline are often in the X and Y position of collagen (Brodsky & Eikenberry, 1982; Kelly, Tanaka, Hardt, Eikenberry, & Brodsky, 1988). So far, hundreds of OI pathogenic variants in COL1A1 and COL1A2 have been recruited in the Osteogenesis Imperfecta Variant Database (https://oi.gene.le.ac.uk). These pathogenic variants include nonsense, missense, frameshift, and splicing. Nonsense and frameshift pathogenic variants generate truncating protein resulting in haploinsufficiency and mild phenotypes (Forlino & Marini, 2000). But the substitution of glycine within the Gly-X-Y triplet domain of the triple helix causes synthesize incorrectly of pro-α chains and structural abnormalities resulting in a dominant-negative effect and severe phenotypes (Bodian et al., 2009; Liu et al., 2007; Zhang et al., 2012).
Osteogenesis imperfecta type I is the mildest form characterized by the blue sclera, near-normal stature, hearing loss, and dentinogenesis imperfecta, but phenotypes are variable between different OI I patients. Osteogenesis imperfecta I is caused by autosomal dominant pathogenic variants in COL1A1 which were related to a quantitative deficiency of structurally normal type I collagen (Willing, Pruchno, & Byers, 1993). COL1A1 (located at position17q21.33, OMIM #120150, HGNC 2197, NCBI reference sequence NM_ 000088.4) contains 51 exons encoding pro-α1 chain of type I collagen.
Splicing variants are less commonly reported than missense/nonsense variant types (Marini et al., 2007; Stenson et al., 2017). Although many COL1A1 splicing pathogenic variants have been described in previous case reports (Peng et al., 2012; Xia et al., 2008; Zhao, Yang, Guo, & Li, 2014), the pathological mechanism was not well studied. Introns removing from pre-mRNA and exons bonding are significantly important in gene expression. These reactions were catalyzed by spliceosome depending on corrected splicing site recognition (Madhani & Guthrie, 1994; Nilsen, 1994). Most introns have a 5′ splice site beginning with GT and a 3′ splice site ending with AG, and some introns have distinct splice site consensus sequences and exhibit either AT-AC termini or AT-AG termini (Patel & Steitz, 2003).
In this study, we identified a novel COL1A1 splicing pathogenic variant c.3814+1G>T with the first nucleotide of intron 48 is changed from G to T which alters the splice site sequence in an OI I patient. Therefore, we aimed to characterize the molecular effect and mechanism of the COL1A1 GT-AG intron pathogenic variant that causes OI I.
2 MATERIALS AND METHODS
2.1 Ethical compliance
This study was approved by the ethics committee of the Second Hospital of Shanxi Medical University.
2.2 Subjects
In this study, a Chinese family with OI I is recruited and all individuals have given the written informed consent. The proband accepted clinical and physical examinations and provided his medical records. Based on all medical records, clinical presentations, and gene diagnostics, the patient was diagnosed as OI I in the hospital.
2.3 Whole-exome sequencing (WES) and variant confirmation
Whole-exome sequencing test was performed on the blood of the proband (III-1). Library preparation and sequencing were performed using Agilent SureSelect Human SureSelect All Exon V5 kit and Illumina HiSeq 2500. The read length at the end of the pair reaches 150 bp and the sequencing depth of the capture region is approximately 100×, sequencing covered also the intron-exon junction. The sequencing results were compared with the human genome reference sequence (UCSC and NCBI database). To identified single nucleotide variants (SNVs), multiple database annotations (such as dbSNP, OMIM, ESP, Clinvar, 1,000 Genomes, and HGMD) were performed simultaneously, and Sanger sequencing was used to confirm the variant.
2.4 Sequencing analysis
The coding region and exon-intron splice junctions including the variant of COL1A1 were analyzed using PCR amplification in combination with Sanger sequencing using the primers (Fw:5′-TCAAGAGAAGGCTCACGATGG-3′; Rv:5′-GGGGTTCTTGCTGATGTACCA-3′). The length of the product is 372bp included exon 48, intron 48, and exon 49. Genomic DNA was extracted from peripheral blood samples and PCR products were purified. These methods were similar to those we used previously (Li, Zhang, et al., 2019).
2.5 qRT-PCR analysis
We used the qRT-PCR to evaluate the transcript variants of COL1A1 in the blood cells from patients and healthy relatives were similar to those we used previously (Li, Zhang, et al., 2019). The primer pair for qRT-PCR of COL1A1 were used: Fw: 5′-GAGAGTACTGGATTGAC-3′ and Rv: 5′-ACTGGAATCCATCGGTCATG-3′. The length of the product is 191 bp including exon 49 of the total (WT + mutated) COL1A1 (NM_000088.4). For normalization, the expression of GAPDH (Fw: CGGAGTCAACGGATTTGGTCGTAT; Rv:AGCCTTCTCCATGGTGGTGAAGAC).
2.6 Prediction of splicing
To assess the presumptive effect of this variant on splicing, three different in silico prediction tools: MaxEntScan (MES) using Maximum entropy score (http://genes.mit.edu/burgelab/maxent/Xmaxentscanscoreseq.html), Human Splicing Finder (HSF, http://www.umd.be/HSF/), and Spliceman (http://fairbrother.biomed.brown.edu/spliceman/) were used.
The output score of each tool indicates the splicing signal strength: the higher the value, the higher the possibility of being a splicing site in vivo. The following equation was used to calculate the percent score variation (%): [(WT score − Mut score)]/WT score × 100 (Miao et al., 2017). The magnitude of the variation corresponds to the increase in the likelihood of mis-splicing. Only the changes greater than 15% (yes/no) were considered to be deleterious significantly (Sharma et al., 2014).
2.7 Cell culture
HEK293T cells were grown in DMEM supplemented with 10% (v/v) FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C and 5% CO2. Transfection was performed using the Polyetherimide (PEI) (PolyScience, Cat.No. 23966-2) according to the manufacturer's instructions. Transfected cells were incubated for 24–48 hr posttransfection.
2.8 The minigene constructs and Western blotting
Minigene constructs encompassing exon 48, intron 48, and exon 49 were amplified using the primer pair of COL1A1 E48-in49-E49 Fw: 5′-GCGCTCGAGGACACCACCCTCAAGAGCCTGA-3′ and E48-in49-E49 Rv: 5′-GCGCTCGAGTACACGCAGGTCTCACCAGTCTC-3′. The amplified minigene products were cloned into the pEGFP-C3 cloning vector at the Xho I and EcoR I sites. The complete sequences of the minigene constructs were validated by Sanger sequencing. Minigene constructs were transiently transfected in HEK293T cells with polyetherimide (PEI) (PolyScience, Cat.No. 23966-2) and RT-PCR was performed with the same primers. RT-PCR products were separated by electrophoresis analysis and isoforms were identified by Sanger sequencing. Besides, we used Western blotting to analyze the proteins of minigene. These methods were similar to those we used previously (Li, Zhang, et al., 2019).
2.9 Analysis of evolutionary conservation of amino acid residues and structure prediction of the mutant protein
NCBI Protein blast was used to analyze the conservation of amino acid residue alteration by comparing across different species. The homology modeling programs Swiss-Model was used to develop an appropriate model to mimic the effects of the mutated region. The structures were displayed by PDB-Viewer software. These methods were similar to those we used previously (Li, Zhang, et al., 2019).
3 RESULTS
3.1 Family pedigree/patient information
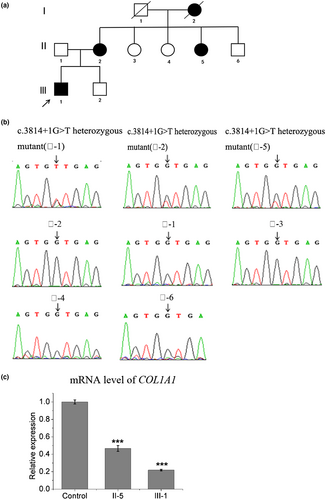
This study included four affected individuals and six unaffected relatives from a Chinese family clinically characterized as type I OI. The pedigree of the family is shown in Figure . The proband (Patient 1, III-1), is a 30-year-old Chinese male, 175 cm tall, and 80 kg in weight. His clinical features include blue sclera, mildly excessive dorsiflexion of fingers, and five fractures. Hearing, teeth, and vision are normal. As he was concerned about his next generation and how this disease was inherited, a directed genetic study and genetic counseling were performed. All patients (II-2, II-5, I-2, and III-1) presented with blue sclerae, DI, and multiple bone fractures. The clinical features of the pedigree are summarized in Table 1. All patients have not accepted bisphosphonates or any other therapies.
| Subject | III-1 | II-2 | II-5 | I-1 (Died) |
|---|---|---|---|---|
| Sex | Male | Female | Female | Female |
| Age (y) | 30 | 51 | 40 | 68 |
| Height (cm) | 175 | 155 | 153 | Unknown |
| Weight (kg) | 80 | 45 | 42 | Unknown |
| Sclerae | Blue | Blue | Blue | Blue |
| Fractures | Five | Six | Multiple | Multiple |
| Dentinogenesis imperfecta | Yes | Yes | Yes | Yes |
| Hearing loss | No | Yes | No | Yes |
| Stature | Normal | Normal | Normal | Unknown |
3.2 Sequencing analysis
Peripheral blood samples were collected from this family including three patients (Ⅱ-2, Ⅱ-5, Ⅲ-1) and five healthy subjects (Ⅱ-1, Ⅱ-3, Ⅱ-4, Ⅱ-6, Ⅲ-2).
Given the proband's requirements, genetic counseling and WES was conducted. After bioinformatics analysis, a novel COL1A1 splicing variant c.3814+1G>T was detected which was confirmed by Sanger sequencing.
To investigate whether this variant cosegregated in this family, Sanger sequencing of genomic DNA isolated from the peripheral blood samples in this family was performed for the COL1A1. The results showed that three patients harbor the heterozygous COL1A1 variant c.3814+1G>T comparing to the five unaffected relatives (Figure 1b). The results revealed that the proband (Ⅲ-1) inherited the described COL1A1 variant (c.3814+1G>T) from his mother (Ⅱ-2) and this variant separated with phenotypes in this family.
To identify the mRNA expression level of COL1A1 in patient samples, qRT-PCR was performed. Total RNA was isolated from the blood samples of the patients (Ⅲ-1 and Ⅱ-5) and health subject (Ⅱ-3) in this family, qRT-PCR was normalized to that of GAPDH. The results showed that the level of COL1A1 mRNA was decreased to 50% (patient: II-5) or 35% (patient: III-1) in the samples from patients compared to control (Figure 1c). These results indicate that the mRNA of mutant COL1A1 is not stable in the cells thus lead to a lower expression of the protein, and implies that an insufficient normal COL1A1 in the patient.
3.3 Prediction of splicing and analysis of evolutionary conservation of amino acid residues
The effect of the c.3814+1G>T variant was computationally analyzed using three splice site prediction programs: MES, HSF, and Spliceman. The outcomes were summarized in Table 2.
| Splicing prediction tool | Prediction score values | ||
|---|---|---|---|
| HSF(%) | MaxEnt scan(%) | Spliceman | |
| COL1A1 c.3814+1G>T | 0.90/0.64 (30) | 6.13/−2.38 (138) | YES:66 |
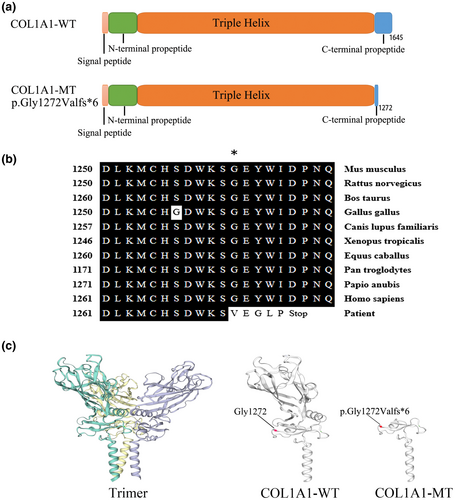
The analysis of amino acid residues showed that these lost amino acid residues were highly evolutionary conserved among COL1A1 proteins from different species (Figure 2b), the mutant protein results in a truncated protein lacking the C-terminal (Figure 2c).
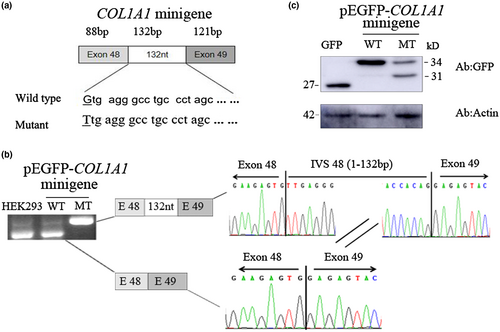
3.4 Splicing analysis of COL1A1 c.3814+1G>T in the minigene
The minigene analysis was performed to identify abnormal splicing. We constructed a minigene containing part of exon 48, exon 49, and entire intron 48 with G or T at c.3814+1 of COL1A1 (Figure 3a). Electrophoresis analysis of RT-PCR products shows that no different band size between untransfected and wildtype minigene transfected HEK293T cells which were identified as COL1A1 exon 48 (121 bp) and exon 49 (88 bp) in both, but a dissimilar band was detected in mutant minigene transfected cells. Sanger sequencing of RT-PCR products reveals the wild type construct is a normal size band of 209 bp which is consistent with COL1A1 exon 48 (121 bp) and exon 49 (88 bp). The c.3814+1G>T substitution resulted in a band of 341 bp which contains 132 nucleotides from intron 48 (Figure 3b). Third, Western blot analysis was performed to study the protein expression of the minigenes. In pEGFP-C3 vector-transfected HEK293T cells, a strong band was observed with the molecular weight of about 27 kD which represents the GFP protein alone (Figure 3c). In pEGFP-COL1A1-minigene wild type transfected cells, a band with a molecular weight at ~34 kD was detected which was identified as the protein products including part of exon 48 and exon 49 fusion with GFP (Figure 3c). But, in pEGFP-COL1A1 minigene mutant transfected cells, one extra band at ~31 kD molecular weight was observed compared to wild type COL1A1 minigene transfected cells, suggesting that the c.3814+1G>T substitution caused a frameshift resulting in early termination of protein expression which is corresponding to the results of alternative transcript analysis (Figure 3c). According to the American College of Medical Genetics and Genomics (ACMG) guidelines for interpretation of sequence variants (Richards et al., 2015), we consider this variant to be pathogenic (PM2 + PVS1+PP1 + PP4).
4 DISCUSSION
Pathogenic variants in type I collagen-encoded genes (COL1A1 and COL1A2) or other genes (CRTAP: MIM#605497, WNT1: MIM#164820, and BMP1: MIM#112264 et, al) responsible for OI. Osteogenesis imperfecta patients can be confirmed and new pathogenic mutations can be discovered by the genetic tests which are significantly important for us to analyze the relationship between the genotype and phenotype of OI comprehensively. Whole-exome sequencing is a currently available effective approach for screening pathogenic variants of OI (Mackenroth et al., 2016). We use WES to detect a pathogenic gene variant which confirmed by Sanger sequencing and perform a series of molecular biology methods to study the pathogenic mechanism of the variant. A novel splicing variant (NM_ 000088.4: c.3814+1G>T; NP_000079.2: p.Gly1272Valfs*6) in COL1A1 was identified in this Chinese family with OI I. Three patients (Ⅱ-2, Ⅱ-5, and Ⅲ-1) carried the novel splicing variant c.3814+1G>T. Five healthy family members (Ⅱ-1, Ⅱ-3, Ⅱ-4, Ⅱ-6, and Ⅲ-2) were free of this variant. These results indicate that the novel splicing variant c.3814+1G>T is cosegregated with the disease in the pedigree.
Given the variant is located at the boundary of exon 48 and intron 48, and alternative splicing depended on the flanking intronic regions recognized correctly (Patel & Steitz, 2003). Owing to pathogenic variants of splice sites could have led to splicing defects (Scotti & Swanson, 2016). To further investigate whether this mutation affects the alternative splicing or not, minigene was performed to mimic the process of splicing in vitro. The minigene results suggest that the novel splicing variant c.3814+1G>T contributed to the disappearance of the 5′ donor splice site of intron 48, which leads to intron 48 retention. This in-frame insertion created a premature termination stop codon (PTC). Mutations that created premature termination codons (PTCs) can cause haploinsufficiency via nonsense-medicated decay (NMD) (Maquat, 1995).
It has been reported that variants at splicing sites can cause OI (Schleit et al., 2015; Schwarze, Starman, & Byers, 1999). These variants included the typical splicing variants which were located at splicing donor/acceptor sites in introns (Schleit et al., 2015), and the atypical splicing sites beyond the splicing sites (GT-AG) (Li, Cao, et al., 2019). A previous study identified a COL1A1 C-propeptide pathogenic variant (one out-of-frame splicing error), generating a PTC and causing haploinsufficiency via nonsense-medicated decay (NMD), was associated with OI I (Symoens et al., 2014). Synthesizing of single COL1A1 allele results in haploinsufficiency which involves nonsense-mediated mRNA decay or frameshift/splicing mutation-induced pretermination codon, most of them being mild OI type (Rauch, Lalic, Roughley, & Glorieux, 2010). In our case, the COL1A1 novel splicing variant c.3814+1G>T (p.Gly1272Valfs*6) is located at the C-terminal propeptide and generated a premature termination codon (PTC). The level of COL1A1 mRNA was decreased in patients (III-1, II-5), so this PTC leads to nonsense-mediated mRNA decay (NMD), by which mRNAs are degraded before they produce large quantities of truncated proteins. The NMD process contributes to the COL1A1 haploinsufficiency which may be the major mechanism underlying the OI I in this family.
In conclusion, we report a novel COL1A1 intronic pathogenic variant c.3814+1G>T alters the alternative splice of COL1A1 and consequently caused Type OI I. Our study validates that c.3814+1G>T is a type OI I-causative mutation and data in this study enrich mutation OI database and provide an OI causative mutation for molecular diagnosis as well as scientific information.
ACKNOWLEDGMENTS
We thank the proband and his family members for their support and participation.
CONFLICT OF INTEREST
Yaxin Han, Dongming Wang, Jinli Guo, Qiuhong Xiong, Ping Li, Yong-An Zhou and Bin Zhao declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
YH wrote the manuscript and performed the practical work. DW collected patient's data. QX and GJ analyzed the patient's data and were assisted by DW. PL designed the study. BZ and YA-Z conceived the study and edited the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.