Interpretation challenges of novel dual-class missense and splice-impacting variant in POLR3A-related late-onset hereditary spastic ataxia
Abstract
Background
RNA polymerase III (Pol III)-related disorders are autosomal recessive neurodegenerative disorders caused by variants in POLR3A or POLR3B. Recently, a novel phenotype of adult-onset spastic ataxia was identified in individuals with the c.1909+22G>A POLR3A variant in compound heterozygosity.
Methods
Whole-exome sequencing was performed in the proband and parents. Variants were confirmed by Sanger sequencing. RNA sequencing was performed to evaluate splicing implications.
Results
A 42-year-old female was evaluated for unexplained neurological findings with a slow progressive decline in gait and walking speed since adolescence. WES revealed a novel missense variant (c.3593A>C, p.Lys1198Arg) in exon 27 of POLR3A in compound heterozygosity with the c.1909+22G>A variant. Summary of previously reported clinical features from individuals with pathogenic biallelic alterations in POLR3A and adult-onset phenotype is consistent with our findings. RNA analysis revealed c.3593A>G drives the production of four RNA transcript products each with different functional impacts.
Conclusion
The novel dual-class c.3593A>C variant in POLR3A causes an amino acid substitution and complex disruption of splicing. Our report supports the need to investigate variants near splice junctions for proper interpretation. Current interpretation guidelines need to address best practices for inclusion of predicted or measured transcriptional disruption pending functional activity or reliable transcript abundance estimates.
1 INTRODUCTION
POLR3A-related neurodegenerative disorders are primarily caused by pathogenic variants in POLR3A and POLR3B (and interacting partners POLR1C and POLR3K) with onset ranging from the neonatal period to adulthood (Bernard et al., 2011; Wambach et al., 2018). POLR3A and POLR3B are the largest subunits of the RNA polymerase III (Pol III) crucial for synthesis of small RNAs, such as 5srRNA and transfer RNAs (tRNAs) (Arimbasseri & Maraia, 2016). Variants in POLR3A are known to cause hypomyelinating leukodystrophy in childhood (POLR3A-HLD) with cerebellar symptoms, with or without hypodontia and hypogonadotropic hypogonadism (sometimes referred to as 4H syndrome; MIM: 607694; Bernard et al., 2011). POLR3A biallelic defects have also been shown to cause Wiedemann–Rautenstrauch Syndrome (WRS; Paolacci et al., 2018; Wambach et al., 2018), a neonatal-onset progeria (MIM: 264090).
A milder and late-onset phenotype of spastic cerebellar ataxia was initially reported in three individuals (Azmanov et al., 2016) with intronic homozygous variants. Later, 18 individuals were described with an intronic hypomorphic allele (c.1909+22G>A, rs191875469), which activates a cryptic donor site and leads to an out-of-frame product in compound heterozygosity with other pathogenic variants (Minnerop et al., 2017). Additional compound heterozygotes harboring the c.1909+22G>A were described in Norway (nine families; Rydning et al., 2019) and Spain (six families; Infante et al., 2019) with a consistent clinical picture of variable-onset ataxia with or without tremor, and a potentially pathognomonic abnormality in the superior cerebellar peduncles (SCP).
To date, there is no designation of this strong genotype-phenotype association in OMIM (MIM:614258). ClinVar submissions exist with conflicting interpretations, and pathogenicity of c.1909+22G>A is a matter of debate. We report a North-American individual with adult-onset spastic ataxia and bi-allelic variants in the POLR3A gene identified by whole exome sequencing (WES). Blood RNA sequencing (RNAseq) revealed a novel dual-class (both missense and splice-site) variant in POLR3A generating multiple transcripts with different impacts.
2 MATERIALS AND METHODS
Proband and parents were consented following Mayo Clinic Institutional Review Board approval. Trio blood samples underwent research WES. Proband samples underwent RNAseq. Methodology is described in Appendix S1. Findings underwent clinical Sanger sequencing confirmation by a reference laboratory (GeneDx).
3 RESULTS
3.1 Clinical report
A 42-year-old female was referred to the Mayo Clinic Department of Clinical Genomics and the Center for Individualized Medicine for longstanding unexplained neurological findings. Neurodevelopment was normal, despite difficulty with activities, and waddling gait. At age 18, her condition started to deteriorate with a slow progressive gait decline. Tremors started in the right upper extremity in her third decade, spreading to the left side and the neck.
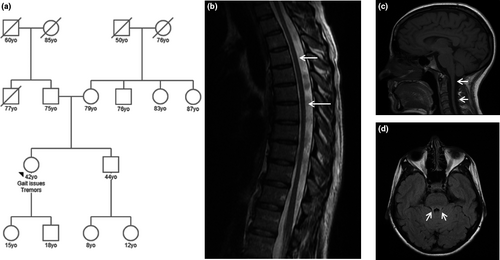
On physical examination, height was normal without dysmorphic features. Cranial nerve (II-XII) examination was unremarkable. Resting and intention head tremor were present. Motor strength was preserved on upper and lower extremities. Deep tendon reflexes were reduced in upper extremities, brachioradialis, knees, and absent at the ankles. Finger tapping was slow bilaterally. Finger-to-nose, finger-to-finger, and heel-to-shin testing showed moderate bilateral dysmetria, and tremor. The proband's gait was wide-based, spastic, and somewhat unsteady. Family history is non-contributory (Figure 1a).
3.2 Biochemical, electrophysiological, and imaging findings
Biochemical characterization was unremarkable (Appendix S2). Electromyography nerve conduction and visual evoked potential (VEP) testing were normal. Somatosensory evoked potentials (SEPs) were performed in the upper and lower extremities. No reproducible cortical responses were obtained with normal absolute latencies and spinal potentials suggestive of impairment in the central proprioceptive pathway, rostral to the cervical spine. Thoracic spine magnetic resonance imaging (MRI) revealed a diffusely small spinal cord without demyelination features (Figure 1b,c). Brain MRI was initially considered normal; reevaluation following genetic findings found hyperintensities in SCP on T2-FLAIR views (Figure 1d).
3.3 Genetic findings
Clinical panel testing for spinocerebellar ataxia repeat expansion (six genes) and a spastic paraparesis gene panel (51 genes) was uninformative. Research WES identified biallelic alterations in POLR3A (NM_007055.3; Appendix S3 and S4). A heterozygous c.1909+22G>A (maternally inherited) variant in intron 12 and a novel heterozygous missense variant, c.3593A>C, p.(Lys1198Arg; paternally inherited) in the penultimate nucleotide of exon 27 were identified. These variants were confirmed by Sanger sequencing in a clinical laboratory and classified as pathogenic and as a variant of uncertain significance (VUS), respectively. Table 1 shows the overlapping phenotype features of our patient with previously reported compound heterozygote cases with a single allele of c.1909+22G>A (N = 44, Table S1). Phenotypic features of patients with homozygous intronic variants is further outlined in Table S2. The c.3593A>C variant is absent in gnomAD (v2.1; Lek et al., 2016). This nucleotide is highly conserved (phastCons: 1.00 and phyloP: 4.89) across mammalian orthologues. The affected lysine residue occurs in the largest domain of the Pol III subunit; where single-stranded DNA from promoter regions binds. Conservation-based tools such as Polyphen2 and SIFT suggest the lysine to arginine change is tolerated. In contrast, MutationTaster, MCAP, and high CADD Phred Score (24.5) suggested the variant is deleterious. SpliceAI (Jaganathan et al., 2019) predicted significant canonical donor site loss (0.85) and a potential donor splice site gain (0.38), located 46 nucleotides downstream of the variant.
| Phenotype features | c.1909+22G>A compound heterozygous, N = 45 | Case |
|---|---|---|
| Age | 43 yearsa (15–68)b | 42 years |
| Age at onset | 19 yearsa (5–51)b | 18 years |
| Ataxia | 100% (45/45) | Yes |
| Abnormal MEP | 100% (25/25) | np |
| Hypopallestenesia | 98% (41/42) | Yes |
| Thin spinal cord | 97% (30/31) | Yes |
| Abnormal SEP | 94%(29/31) | Yes |
| SCP abnormalities | 93% (28/30) | Yes |
| LL spasticity | 79% (35/44) | Yes |
| Dental abnormalities | 63% (24/38) | No |
| Postural/kinetic tremor UL | 50% (22/44) | No |
| Muscle atrophy | 48% (16/33) | No |
| Dysarthria | 47% (21/44) | No |
| Pes Cavus | 45% (5/11) | Yes |
| Myopia | 33% (14/42) | No |
| Corpus Callosum Thinning | 27% (6/22) | Yes |
| Abnormal VEP | 25% (3/12) | No |
| Dystonia | 22% (10/44) | No |
| Polyneuropathy | 26% (7/36) | No |
| Hypogonadism | 5% (1/19) | No |
- Abbreviations: LL, lower limbs; MEP, motor evoked potential; np, not performed; SCP, superior cerebellar peduncles; SEP, somatosensory evoked potentials; UL, upper limbs; VEP, visual evoked potential testing.
- a Mean value.
- b Values range.
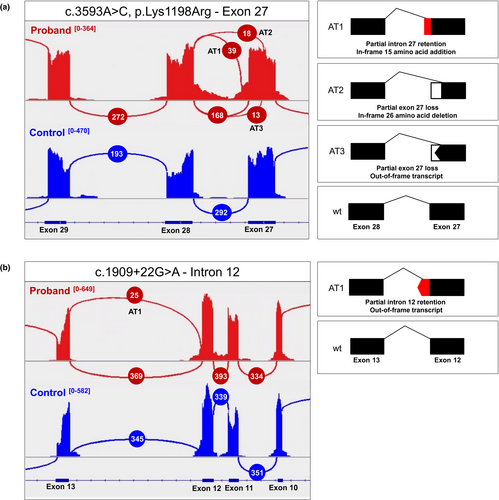
RNAseq from blood revealed c.3593A>G is associated with multiple splicing outcomes (Figure 2a; Appendix S5) without exon skipping (r.=, r.3594_3595ins3594+1_3594+45, 3514_3594del, 3506_3594del]). 70% of junction reads between exons 27 and 28 support canonical splicing (r.=) of which only 2.5% (5/168 reads) contain c.3593A>G. 16.3% of junction reads support use of a cryptic splice donor in intron 27 adding 15 in-frame amino acid residues (r.3594_3595ins3594+1_3594+45, p.Lys1198_Val1199ins(15), Appendix S6) corresponding to the SpliceAI prediction. In addition, 7.5% support use of an exonic splice donor in exon 27 leading to an in-frame 26-amino acid deletion (r.3517_3594del, p.1173Val_1198Lysdel, Appendix S7). Finally, 5.4% support an alternative cryptic splice donor in exon 27 leading to the out-of-frame loss of 89 nucleotides (r.3506_3594del, p.Glu1170Glyfs*16, Appendix S8) predicted to undergo nonsense-mediated decay. Internal control samples (N = 46) from blood RNAseq did not contain any instance of reads supporting the noncanonical junctions observed in the proband. Finally, the c.1909+22G>A allele drove 7% of observed transcripts to use a cryptic intron donor predicted to cause an out-of-frame product similar to previous reports (Figure 2b).
4 DISCUSSION
Here we describe the use of WES to identify a novel POLR3A missense and splice variant, c.3593A>G, in a North American female with adolescent-onset spastic ataxia. The variant is compound heterozygous with c.1909+22G>A, previously observed in similarly affected individuals from European countries. Systematic evaluation of reported phenotypic findings shows significant overlap with our proband. RNAseq demonstrates the c.3593A>G variant produces 4 different splicing outcomes; each with a different functional effect including a missense change with normal splicing, an in-frame insertion, an in-frame deletion, and an out-of-frame deletion. To the best of our knowledge, this is the third case of POLR3A late-onset spastic ataxia phenotype in North America.
Historically, functional characterization of POLR3A variants has proven difficult. Studies on mice generated with homozygous p.Gly672Glu did not recapitulate neurological abnormalities (Choquet et al., 2017). Conversely, homozygous p.Arg103His causes embryonic lethality (Choquet, Pinard, et al., 2019) via defective Pol III complex assembly (Choquet, Pinard, et al., 2019). Gene expression dysregulation by defective POLR3A, specifically of myelin is hypothesized as a potential mechanism for POLR3A-HL disease (Choquet, Forget, et al., 2019). However, evidence is lacking regarding c.1909+22G>A role in triggering late-onset POLR3A disease. Systematic assessment of clinical features in 44 compound heterozygotes carrying the c.1909+22G>A allele highlights a consistent phenotype with mean onset of ataxia near the second decade. Most cases present with spinal cord atrophy, SCP hyperintensities, hypopallesthesia, and abnormal MEP/SEP with lower limb spasticity. Dental abnormalities, dysarthria, muscle atrophy, and upper body tremors are present in half of reported individuals. Infrequent features are corpus callosum abnormalities, visual deficiencies, polyneuropathy, and dystonia. Interestingly, individuals (N = 11) with homozygous intronic alterations exhibit generally an earlier onset of ataxia (first decade of age) and more severe phenotype than those with the c.1909+22G>A in trans with a different pathogenic variant. Interestingly, a Chinese woman with atypical adult-onset 4H syndrome was reported with homozygous c.1909+18G>A with ataxia onset in her third decade (Yang et al., 2019). Hence, intronic variation appears to be an important driver for developing a variable late-onset phenotype.
Clinical suspicion of POLR3A spastic-ataxia and intron sequencing inclusion on commercial panels during genetic workup could benefit our understanding surrounding this phenotype and its prevalence.
In addition, we demonstrate that coupling WES and RNAseq enhances variant interpretation and reveals complex functional consequences. Interpretation following recommendations (Abou Tayoun et al., 2018) for LoF (PVS1) criteria enables the c.3593G>A variant to be conservatively re-classified as Likely Pathogenic. This is an extremely rare variant, previously unreported in populational databases. The described out-of-frame transcript is expected to undergo NMD. The more abundant in-frame insertion and in-frame deletion occur in a DNA-binding region critical for protein function; hypothetically resulting in a loss of protein or enzymatic activity. Transcript differences are confined to an exon represented across all isoforms and a region critical for protein function. Thus, a PVS1_strong level of evidence is applicable (while withholding the application of PM4 and PP3 criteria). However, recommendations and best practices for integration of RNAseq evidence into functional and/or predictive evidence criteria of ACMG/AMP classification system is under evaluation by the ClinGen consortia. The criteria and level of evidence transcriptomic data represents with an uncertain estimate of transcript abundance is currently missing. For example, the third abnormal transcript we describe may be underrepresented, since an out-of-frame product typically undergoes NMD. Furthermore, recommendations for the standarized use of supporting functional in-vivo or in-vitro evidence (and applicable level) if available in parallel to RNAseq or other transcript analysis are not available.
In summary, we describe the emergent late-onset spastic ataxia clinical features in 45 reported compound heterozygotes with c.1909+22G>A, and report a novel single nucleotide variant generating a missense change as well as multiple splicing patterns each with a different functional impact. We provide another example demonstrating the need for comprehensive interpretation guidelines regarding the use of transcript level studies like RNAseq as they become more commonly utilized in clinical and research settings.
ACKNOWLEDGMENTS
This work was supported by the Mayo Clinic – Center for Individualized Medicine (CIM), Translational Omics Program. We would also like to thank the proband and family for their participation in this study.
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose.
AUTHOR CONTRIBUTIONS
JMR and ELM analyzed data and drafted manuscript. JMR, MAC, RD, and GRO contributed to data collection and analysis. RD and EWK supervised the work. All authors read and approved the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
Available from the corresponding author upon reasonable request.