Atomistic MD Simulations of n-Alkanes in a Phospholipid Bilayer: CHARMM36 versus Slipids
Abstract
Linear alkanes (n-alkanes) are chemically the most simple linear hydrophobic molecules in nature. Studying the incorporation of n-alkanes into lipid membranes is therefore a good starting point toward understanding the behavior of hydrophobic molecules in lipid membranes and to assess how accurately molecular dynamics models describe such systems. Here, the miscibility and structure of different n-alkanes—n-decane (C10), n-eicosane (C20), and n-triacontane (C30)—in dipalmitoylphosphatidylcholine membranes are investigated using two of the most used force fields for lipid membrane molecular dynamics simulations (CHARMM36 and Slipids). The n-alkanes are miscible in the membrane up to a critical volume fraction, ϕc, that depends on the force field interaction parameters used. ϕc is dependent on alkane chain length only for the model with more disordered chains (Slipids). Below ϕc, a comparison with 2H nuclear magnetic resonance (NMR) spectra indicates that a more realistic structure of the longer alkane molecules (C20 and C30) is obtained using the Slipids force field. On the other hand, for the shorter alkane (C10), Slipids simulations underestimate molecular order and CHARMM36 simulations enable a precise prediction of its experimental spectrum. The predicted 2H NMR spectra are highly sensitive to 1–4 electrostatic interactions, and suggest that a reduction of the partial charges of the longer alkanes and acyl chains in CHARMM36 results in a better performance. The results presented indicate that lipid membranes with incorporated alkanes are highly valuable systems for the validation of force fields designed to perform lipid membrane simulations.
1 Introduction
Many biological processes related to the incorporation of hydrophobic molecules into cell membranes remain poorly understood at the molecular level. Examples include the anaesthetic effect of alkanes,[1-3] the formation of lipid droplets,[4-6] or the potential effect of microscale and nanoscale plastics on cell membrane structure.[7-10]
One important open question concerns the solubility of hydrophobic molecules in a membrane system. It is yet unclear how the chemistry, size, and shape of hydrophobic molecules affect their structure, dynamics, and partition in a lipid membrane.[11-13] Exploring these dependencies by systematic studies will help to understand their role in biological processes as well as to guide the development of lipid-based technologies.
Linear alkanes (n-alkanes) are the most simple hydrophobic chain-like molecules in nature for which extensive data has been collected. Therefore, n-alkanes are a suitable starting point to address the problems introduced above. The solubility of n-alkanes in many different solvents has been determined as a function of alkane chain length[14-16] as well as the enthalpy of mixing of various n-alkane binary mixtures.[17] The available data shows that at higher temperatures, the enthalpy of mixing of liquid n-alkanes with different chain length becomes negative, that is, there is complete miscibility.[18, 19] The solubility of alkanes in phospholipid membranes, pertinent to the biophysical questions outlined above, is however more intricate since the lipid bilayer hydrophobic region is a nm-thick fluid possessing interfacial surface tension and a gradient of molecular order in the direction of the bilayer normal.[20-22] Depending on alkane chain length and concentration, the complete mixing of alkanes with the phospholipid acyl chains may imply a significant change of both the surface area and the molecular order of the system. Analogous to this problem, it is well known that the isotropic-to-nematic transition of a nematogenic solvent with dispersed flexible chains (including n-alkanes) induces phase separation, and that the concentration of the flexible chains in the nematic phase decreases with chain length.[23-25] In contrast to nematogens, the acyl chains in a lipid bilayer have a high degree of flexibility. Nevertheless, a similar dependence on n-alkane chain length may emerge due to the fact that longer alkanes have a stronger tendency to fold[26] and thus perturb the lipid membrane to a larger degree: Increased chain folding is expected to lead to a larger increase in local interfacial area-per-phospholipid (APL) and therefore a higher increase of interfacial free energy.
The solubility of alkanes with different chain lengths in lipid bilayers has been investigated experimentally by different research groups with apparently contradicting results. Previous 2H nuclear magnetic resonance (NMR) results reported by Pope et al.,[11, 27, 28] indicate that n-alkanes longer than the lipid acyl chains become immiscible. In contrast, we have recently demonstrated that n-eicosane (C20) and n-triacontane (C30), with chain lengths of 20 and 30 carbons, respectively, are miscible in dipalmitoylphosphatidylcholine (DPPC) membranes (acyl chain length of 16 carbons).[29] The origin of these contrasting observations is very likely the distinct sample preparation procedure used, which we have observed to heavily influence the time needed to attain thermodynamical equilibrium. In the previous work, we showed that with a standard mixing procedure [30]—dissolution of n-alkane and phospholipid in organic solvent, subsequent solvent evaporation, and hydration of the resulting lipid film—it is possible to mix C30 with DPPC lipid membranes up to an alkane-to-acyl chain volume ratio of ≈5%.[29] This is the longest n-alkane that has been directly observed in a lipid membrane to date. From our previous experimental results, it was apparent that the solubility of C30 in DPPC lipid membranes is significantly lower compared to the solubility of C20. However we cannot exclude an effect of the organic solvent used, that is, phase separation may occur during the evaporation of the organic solvent, due to the distinct solubilities of alkanes[15] and phosphatidylcholines,[31] leading to a lipid/alkane film with an inhomogeneous distribution of the two components.
Molecular dynamics (MD) simulations of alkanes in lipid membranes can be used to circumvent such sample preparation shortcomings since the initial arrangement of molecules is fully controlled. In a recent work, we used the CHARMM36 force field (ff)[32] for modeling DPPC bilayers containing n-alkanes, assuming equivalent interactions for both n-alkanes and acyl chains.[29] We had chosen CHARMM36 because of its generally good agreement with experimental NMR order parameters of phospholipids.[33] Such a description led to the observation of a critical n-alkane volume fraction ϕc, at which the bilayer APL reaches a maximum value, and above which alkane chains start to accumulate between the bilayer leaflets.[29] ϕc was identical for 10 and 20 carbon long n-alkanes in contrast to their experimental apparent solubility. Comparison of these simulations with 2H NMR experimental results however showed that, while the CHARMM36 ff is suitable to model n-decane in a DPPC membrane, it significantly overestimates the ordering of longer n-alkanes (C20 or C30).[29] Ideally, to investigate the dependence of ϕc on longer alkane chain lengths, one should therefore use a more realistic model for these chains.
Another ff specifically designed to study lipid membranes is the Slipids ff.[34, 35] The comparison of phospholipid order parameters with experimental data shows that the phospholipid headgroup structure is not as realistic as in CHARMM36.[33, 36] However, since the Slipids ff was developed with particular focus on lipid acyl chains by using a parametrization based on DFT calculations on hexadecane (partial charges), high level ab initio calculations on octane (dihedral torsions) and a comparison with thermodynamical and dynamical properties of bulk alkanes, it is a suitable alternative to CHARMM36 for investigating the behavior of n-alkanes in phospholipid bilayers.
In this work, we perform a systematic investigation of alkanes with different chain lengths—n-decane (C10), n-eicosane (C20) and n-triacontane (C30)—incorporated into DPPC membranes, using both the CHARMM36 and the Slipids force fields. The simulations show that the Slipids ff gives indeed, as expected, more realistic estimates for C–H bond order parameters of the longer n-alkanes due to a higher configurational entropy of the chains. The main contribution for the different performance of the two ffs is the strength of the electrostatic 1–4 interactions and we demonstrate that by reducing the partial charges of the acyl chains in CHARMM36, a better agreement with the 2H NMR spectra of C20 and C30 can be achieved. The dependence of ϕc on alkane chain length is also investigated for both force fields. ϕc is indeed dependent on alkane chain length but not as strongly as apparent from our previous experimental measurements. We believe that the results shown will motivate new experiments (exploring the use of different organic solvents) and should be taken as a valuable reference for future force field optimizations.
2 Results and Discussion
2.1 Slipids Simulations Predict the Structure of the Longer Alkanes More Accurately
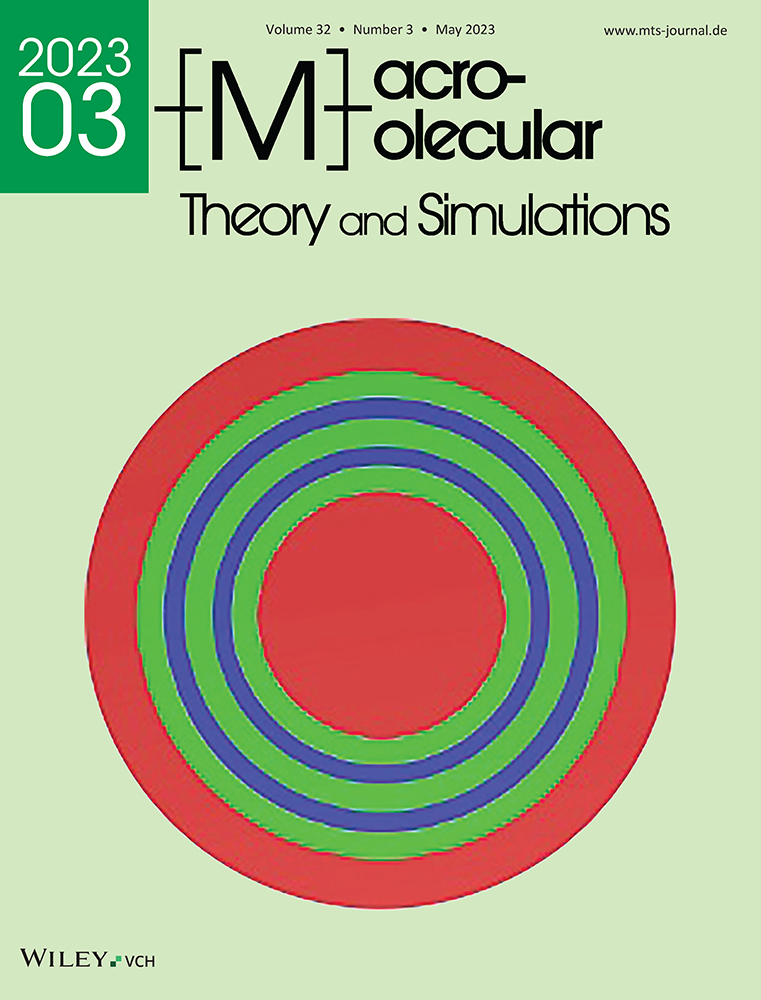
In order to determine if the Slipids force field results in a more realistic structural behavior of the long n-alkane chains than CHARMM36, we simulated three n-alkanes with different chain length in a DPPC bilayer and calculated the alkane C–H bond order parameters. The comparison with experimental results is shown in Figure 1. For reference, we also present the phospholipid SCH magnitudes of a pure DPPC membrane (Figure 1B), together with experimental values from our group and from Seelig et al.[37]
These profiles confirm that the CHARMM36 ff reproduces the phospholipid headgroup order parameters well. However, the absolute acyl tail order parameters are somewhat larger than the experimental values. This observation agrees with previous findings,[39] and is also indicated in the initial CHARMM36 publication:[32] While Klauda et al. achieve very good agreement with the 2H NMR order parameters using a fixed APL of 0.64 nm2 (NPAT ensemble, T = 50° C), their NPT simulation results in a slightly decreased area-per-lipid (62.9±0.3 nm2), and consequently higher ordering of DPPC acyl chains. We note here that our order parameters were consistently larger than those obtained by Klauda et al.[32] (see Figure S1, Supporting Information). This is expected as we are using a simulation software different from the one used by Klauda et al. (GROMACS[40] vs CHARMM[41]) as shown previously,[42] possibly due to different switching functions implemented in the different programs. Recently, a particle-mesh Ewald algorithm has been implemented for the treatment of long-range dispersion eliminating the dependence on the cut-off switching function and is expected to improve the accuracy of lipid acyl chain order parameters[43, 44] and liquid alkane properties.[45] However, we find that the alkane order parameters remain largely unaffected by slight variations of the simulation settings, or even the lipid order (see below), and thus we simply used the default simulation settings provided by the CHARMM-GUI membrane builder.
Simulations with the Slipids ff result in a better agreement with the experimental DPPC acyl chain order parameters as expected,[34, 39] but far worse order parameters for the headgroup and glycerol carbons (compared to CHARMM36), as also shown previously.[33] We note here that the updated (2020) version of the Slipids ff addresses the problem of the glycerol carbon order parameters.[35] However, we did not use that version as we are focusing solely on the hydrocarbon chains in this work. Furthermore, a test on the DPPC/n-alkane systems with the 2020 ff version yielded no improvement with regards to matching the experimental n-alkane order parameters.
With respect to the n-alkane order parameters, both CHARMM36 and Slipids only produce satisfactory results for certain alkane lengths. This is shown in Figure 1C, which compares simulated 2H NMR spectra (calculated from the corresponding order parameter profiles) with experimental spectra from deuterated alkanes (C10d, C20d, and C30d). The manual fitting procedure used to obtain the experimental order parameter profiles is outlined in Figure S2, Supporting Information. The measurements were conducted at the same conditions (hydration, alkane volume fraction, and temperature) as the simulations. For C10, the spectrum predicted by the CHARMM36 simulation matches the experimental spectrum nearly perfectly, while the Slipids order parameters are closer to zero, resulting in a spectrum which is much too narrow. For C20 and C30 on the other hand, the quadrupolar splittings determined from the Slipids simulations match the experimental result well, while CHARMM36 significantly overestimates these splittings.
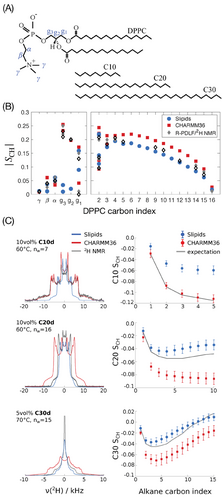
The observed chain-length dependent performance of the two force fields can be understood by looking more closely at the n-alkane order parameter profiles in Figure 1C. The maxima of the order parameter in the middle of the C20 and C30 chains are notably higher in Slipids than in CHARMM36 simulations. These maxima indicate that the alkane chains have a more disordered character and therefore fold more easily when using the Slipids ff compared to CHARMM36. The higher conformational freedom is confirmed by the consistently smaller trans-gauche ratio of the n-alkane carbon dihedrals in Slipids simulations, as shown in Figure 2 . Similar observations have been made by Hötgen et al.,[46] who point out that gauche conformations are under-represented in the CHARMM27 ff, a predecessor of CHARMM36.
We have also performed simulations of neat alkanes to understand how the trans-gauche ratios of the alkane chains vary from bulk to the lipid bilayer and for experimental comparisons. Experimentally it has been determined previously by Mengers and D'Angelo that the 3JCC of neat n-undecane at 25 °C in various solvents was equal to 3.6±0.1 Hz which translates to a trans-gauche ratio of 3.17 using a Karplus equation.[47] On the other hand, Casal et al. show that about 3.5 gauche conformers can be expected in a tridecane molecule at 30 °C, which equals a trans-gauche ratio of ≈2.4.[48] This value is similar to the trans-gauche ratios we obtained for bulk decane in CHARMM36 simulations, at the same temperature (see Figure S3, Supporting Information), but probably underestimates the amount of gauche conformers, according to the authors. Using CHARMM27, Klauda et al. previously calculated trans-gauche ratios of ≈2.0–2.5 for heptane,[49] while Thomas et al. observed higher amounts of trans conformers in Monte Carlo simulations of n-alkanes up to 12 carbons in length (trans-gauche ratios of ≈4.0).[50]
In our simulations, the trans-gauche ratio of alkane molecules calculated from bulk alkane systems is nearly the same as in the lipid bilayers, as shown in Figure 2. In addition, these trans-gauche are similar to those calculated for the lipid acyl tails, toward the end of the tail. This is in line with the previous observation that the phospholipid acyl tails in a lipid bilayer have a liquid like behavior very close to the behavior in bulk, as observed from experimental NMR spin-lattice relaxation rates and MD simulations.[51] We can thus conclude that the non-zero n-alkane order parameters of alkanes within these lipid membrane models are mainly a result of overall anisotropic orientation, rather than a combination of orientational and conformational effects.
We note that the simulations and experiments for C10, C20, and C30 were conducted at different conditions, such as a very low level of hydration for the C10 system. While the CHARMM36 ff was tested and shown to provide good results at such low water contents,[32, 33, 36] the Slipids ff was not yet tested in this context (to our knowledge). One could therefore suspect that the Slipids order parameters of C10 would agree better with the experiment at higher water contents. However, this is not the case, as shown in Figure S4, Supporting Information.
2.2 Reducing the Partial Charges of Alkyl Chains in CHARMM36 Enables to Reproduce the 2H NMR Spectra of Long Alkanes
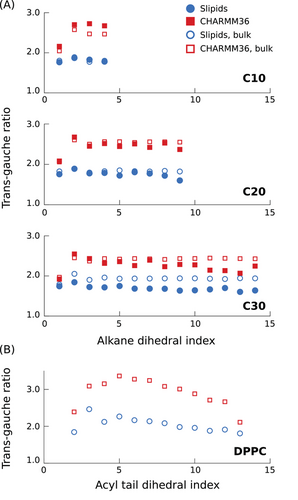
In this section we explore why the n-alkane chains behave differently in the CHARMM36 and Slipids simulations. The SCH profile of saturated acyl chains is quantitatively related to the bilayer surface area as described in detail by Petrache et al.[52] An increase in surface area leads to more disordered acyl chains. Therefore, our initial hypothesis was that the reduction of n-alkane order parameters in Slipids simulations is a consequence of the larger area per lipid obtained for the Slipids ff in comparison to CHARMM36.[39] To test this hypothesis, we have performed NPAT simulations at fixed surface areas per phospholipid, equal to the average interfacial APL molecule, 〈A〉, determined from the simulations in the NPT for each force field. The results are shown in Figure S5, Supporting Information. By performing CHARMM36 simulations at a constant surface APL corresponding to the value determined from the Slipids simulations, 〈A〉SL, a reduction of both the lipid and alkane order parameters was indeed observed. However, for the alkane, the reduced (absolute) order parameters were still notably larger than those obtained from NPT Slipids simulations. In addition, alkane ordering in Slipids simualtions was not significantly affected by the change in APL. This observation indicates that the reduced ordering of n-alkane molecules in the Slipids ff is a consequence of intramolecular interactions, rather than due to the different interfacial free energy minima of the bilayers.
To further investigate the main source of the differences between the two force fields, we generated hybrid force fields to describe acyl chains and alkanes by systematically replacing some of the force field parameters of Slipids by parameters from CHARMM36 and observe the effects on the alkane SCH values. The results are shown as Supporting Information (Figure S6, Supporting Information) and can be summarized as follows: If the Slipids scaling factors for the 1–4 Coulombic interactions and for the 1–4 van der Waals (vdW) interactions are kept unchanged (0.8333 and 0.5, respectively), nearly no changes are observed in the SCH profiles when substituting the Slipids ff parameters that describe the phospholipid acyl chains and alkanes with the parameters from CHARMM36, namely the parameters for dihedral torsions, Lennard–Jones (LJ) interactions, and partial charges. Nevertheless, in such conditions, certain trends can be detected. The substitutions of the partial charges (from zero to non-zero values in CHARMM36) and vdW parameters lead to an increase in alkane order, while the substitution of the dihedral torsion potentials has the opposite effect. If all these parameters are changed simultaneously there is no meaningful change of alkane order. In contrast, if in addition to the use of the CHARMM36 partial charges one takes the scaling factor for the 1–4 Coulombic interactions from CHARMM36 into account, the increase of alkane order is greatly amplified, leading to SCH magnitudes higher than for CHARMM36. This observation enables us to pinpoint the stronger 1–4 electrostatic interactions as the source of the higher order in CHARMM36. This result is in line with previous findings.[46] We have therefore performed CHARMM36 simulations with a reduction of partial charges for the alkanes and acyl chains. The results are shown in Figure 3. This reduction of partial charges results in both the long alkane and the acyl chain order parameter profiles becoming closer to the experimental ones. This suggests that by scaling the set of partial charges, it is possible to tune the force-field to yield a nearly perfect match between the simulated and the measured alkane 2H NMR spectra. We refrain from doing this here.
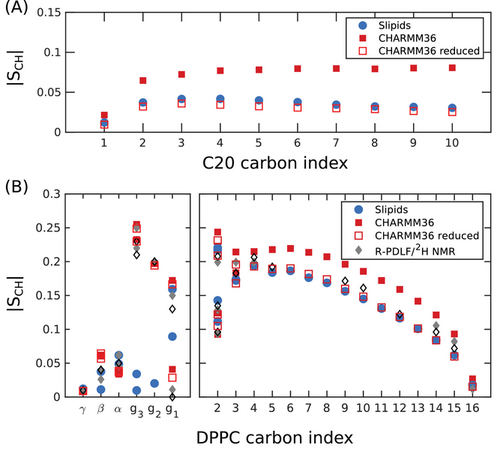
It is not clear to us how the partial charges for acyl chains in the CHARMM36 ff were defined originally. They can be traced down as far as to the work by Feller and Mackerell in 2000,[53] but very likely they have been already defined in the first atomistic CHARMM force field in the early 1990s.[54] The Slipids force field uses partial charges averaged from DFT calculations performed on the most probable alkane structures from an iterative procedure using bulk MD simulations.[34] It is possible, as indicated by the results shown here, that a reduction of the partial charges in CHARMM36 leads to more realistic models. Such a hypothesis needs to be assessed by means of experimental observables other than the C–H bond order parameters. For instance, the validation of dynamics time-scales can be done by measuring NMR relaxation rates from these systems to enable comparisons with the MD predictions, as demonstrated previously.[32, 36, 55] Nevertheless, an important remark is that here the use of the alkane order parameters as target experimental observables (i.e., the fitting target) for tuning the CHARMM36 force field leads to a better match also for the DPPC acyl chain order parameters (which were not taken into account for the optimization) as can be seen in Figure 3B. The parametrizations of common lipid force fields have mostly been done by considering structural and thermodynamical observables from single component systems, or binary systems containing cholesterol.[56, 57] We believe that for future ff optimizations, the use of experimental data from alkane/phospholipid mixtures should be taken as a valuable assessment tool, especially considering the chemical equivalence of alkanes to acyl chains, and the highly entropic (positional and conformational) nature of alkanes in a lipid bilayer in contrast to the rigid nature of cholesterol molecules.
2.3 The Effect of Alkane Chain Length on Alkane-Lipid Miscibility Depends on the Force Field Used
In order to investigate the effects of the alkanes on simple properties of the model lipid bilayers, and how such properties depend on chain length and interaction parameters, we conducted additional MD simulations at identical (NPT) conditions for the three n-alkanes for a range of alkane volume fractions (0–30 vol% of n-alkane each). We simulated two groups of systems, one group in a low hydration regime (nw = 7) and a second group with a number of water molecules between bilayers closer to the excess water limit (nw = 19). Figure 4 shows the variation of bilayer thickness and APL on alkane volume fraction for all simulations. Every plot also shows the calculated values for the average quantity corresponding to the bilayer without alkane. From these values it is clear that by using the CHARMM36 interaction parameters, one predicts more ordered membranes (lower average area and higher thickness) than Slipids. As mentioned above, using the LJ-PME option could improve the accuracy of the APL obtained from CHARMM36 simulations. Rather than trying to use the optimal settings for CHARMM36 simulations, we here choose to use identical simulation settings for both the CHARMM36 and Slipids interaction sets, such that the differences observed between models depend only on interaction parameters. The simulation settings used for the CHARMM36 simulations were therefore the same as the settings described in Table 1 for Slipids. No significant differences for the order parameters obtained with these settings have been found as shown in Figure S1, Supporting Information.
| Programs | CHARMM36 | Slipids |
|---|---|---|
| Cutoff-scheme | Verlet | Verlet |
| vdw_modifier | Force-switch | None |
| rvdw [nm] | 1.2 | 1.2 |
| rvdw_switch [nm] | 1.0 | - |
| coulombtype | PME | PME |
| rcoulomb [nm] | 1.2 | 1.4 |
| tcoupl | Nose–Hoover | v-rescale |
| tau_t [ps] | 1.0 | 0.1 |
| pcoupl | Parrinello–Rahman | Parrinello–Rahman |
| tau_p [ps] | 5.0 | 1.0 |
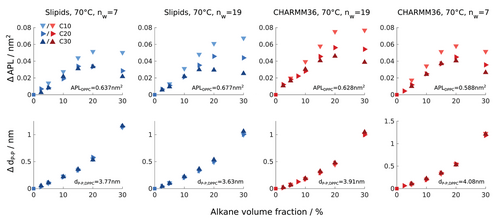
The bilayer thickness, here defined as the average distance between the phosphor planes of the two leaflets, increases in nearly exactly the same manner for the three n-alkanes, irrespective of the set of interaction parameters used and of the hydration state of the system. Generally, the thickness increase per added alkane becomes larger with increasing alkane content. This is in line with our previous results using the CHARMM36 ff.[29] The APL increases with alkane concentration for both hydration levels and force fields toward a maximum value that we define as ϕc. This observation is similar to what we found previously, and signals that the alkane molecules partition primarily into the acyl chain region below ϕc, while at higher concentrations the molecules accumulate more and more in the bilayer center.[29] At ϕc, the maximum APL is largest for C10 and smallest for C30 for both force fields. Taking the thickness dependence into account, this means that the bilayers with C10 and C30 have the lowest and highest density, respectively. For simulations with CHARMM36, ϕc is independent of chain length and is ≈20 vol%. For simulations with Slipids, and at higher hydration, ϕc depends on alkane chain length with ϕc between 20 vol% and 30 vol% for C10, ϕc ≈20 vol% for C20, and to 15 vol% for C30. At lower hydration, the differences of ϕc for the distinct chain lengths obtained with Slipids are attenuated. Above ϕc, particularly for long alkanes and low water contents, the APL drops from 20 to 30 vol% of alkane.
Due to the pure hydrophobic nature of the alkanes, an increase of local area leads to more hydrophobic contacts between water molecules and the hydrophobic region. At ϕc, addition of decane results in the largest areas, that is, at the same volume fraction, smaller chains induce a higher increase of interfacial area. As the alkane gets longer, bulky U-shape conformations will occur that induce a higher fluctuation of the local area. Assuming a simple parabolic interfacial free energy,[20] the mixing of bulkier chains will therefore lead to a higher penalty of free energy which may disfavor such bulky conformations, leading to a lower average area for membranes with C30. The dependence of ϕc on chain length for the Slipids force field, in contrast to the results from CHARMM36, might also be directly related to the more disordered character of the alkane chains. The chains in Slipids have a higher propensity to fold, as shown in Figures 1 and 2, and this should lead to a stronger increase of local interfacial APL when the chain is located between acyl chains. The effect is however smaller than what is expected from experiments (ϕc ≈ 4–5 vol% for C30[29]). Nevertheless, such dependence of ϕc on alkane chain length is qualitatively predicted by the Slipids model. ϕc values calculated for C20 from both force fields are notably close to the apparent values from our previous experiments.[29]
ϕc should mainly result from the increase of interfacial free energy. If we assume the apparent experimental solubility of hydrophobic molecules in a lipid bilayer to be directly related to ϕc, the comparison of ϕc with solubility for different hydrophobic molecules becomes a highly valuable observable for validation of lipid membrane simulations. We must note here again that it is not clear to us if the experimental values for solubility that we previously measured depend on the sample preparation procedure. New experiments using a range of different sample preparation methods (e.g., different mixing temperature and organic solvents) need to be performed in this context. Several observables have been used to validate force fields for describing bulk alkanes[49, 58-60] and phospholipid bilayers.[39, 49, 53, 54, 61] If the assumption that the apparent solubility observed in NMR experiments and ϕc are the same is indeed correct, a comparison of ϕc with solubility measurements would put the delicate balance of interactions that determines the surface tension described by a force field also to the test, as well as the transferability of interaction parameters from alkanes to acyl chains and vice-versa.
The description presented here concerns the behavior observed from computational models assuming an equivalent representation of the alkane chains irrespectively of their length. This seems to be a reasonable assumption for the longer n-alkanes such as C20 and C30, based on the C–D order parameter profiles calculated from the Slipids simulations which match well with all the experimental NMR values determined so far. Slipids however fails to describe the experimental order parameters of decane molecules which indicates that the assumption for equivalent representations of the alkane chains, starting from the shortest alkanes, may not be possible. This is in line with previous results from computational studies on bulk alkanes.[62]
3 Conclusions
In this manuscript, we compared two state-of-the-art lipid force fields with respect to their ability to describe n-alkanes of different lengths inside a lipid bilayer environment at different concentrations. Using 2H NMR order parameters as the main validation observable, we show that the force field performance varies significantly with alkane chain length. CHARMM36 modeled only small concentrations of C10 in DPPC accurately, while Slipids reproduced the experimental order parameters of C20 and C30 well. By selectively modifying individual ff parameters, we identify the electrostatic 1–4 interactions as the main factor influencing alkyl chain ordering, at least within the parameter ranges used by the CHARMM36 and Slipids ffs. This is in line with previous observations.[46, 62] In the Slipids ff, these 1–4 interactions are virtually absent since the partial charges of most carbons are zero, resulting in more flexible chains compared to CHARMM36. Because of the more disordered character of the alkyl chains in Slipids, the 2H NMR data is well predicted for the longer alkanes with this force field. However, we also show that the CHARMM36 ff can be adapted to give order parameters similar to Slipids simulations, and to predict the 2H NMR measurements, by reducing the partial charges of the alkanes and lipid acyl chains.
The two force fields predict a distinct dependence on alkane chain length for ϕc, the volume fraction at which the APL reaches a maximum with alkane incorporation. ϕc should be a consequence of the interfacial free energy and therefore of the delicate balance between the interactions modeled, and defines the solubility of alkanes in the acyl chain region. We believe that the combination of a large set of lipid membrane simulations, with a variety of alkane/phospholipid mixtures, with the measurement of experimental observables on the corresponding systems (alkane and phospholipid order parameters, X-ray scattering form factors, solubility measurements) would be an invaluable toolkit for force field optimization. The results shown here, particularly for the long alkanes are very encouraging.
The results and discussion presented here should be of general interest for the optimization of MD simulations of lipid membranes, and in particular for future investigations of hydrophobic macromolecules inside lipid bilayers, as well as for the re-emerging interest on the behavior of triglycerides inside lipid membranes in connection to lipid droplet formation in cellular membranes.
4 Experimental Section
Simulation Setup
Atomistic molecular dynamics simulations were performed using the GROMACS software[40] (version 2018.7). The investigated systems consisted of DPPC bilayers containing either C10, C20, or C30. The compositions used in this work are described in detail below and in Table S1, Supporting Information. Most DPPC/C10 and DPPC/C20 topologies were reused from a previous publication.[29] For the remaining systems the setup proceeded accordingly: the initial DPPC topology was downloaded from open access data made available by the NMRlipids project (nmrlipids.blogspot.fi)[63] and equilibrated. Alkane topologies were designed using Molden,[64] equilibrated in bulk, and the necessary number of alkane molecules was placed between the two bilayer leaflets. Then the mixtures were hydrated to the desired amount of water and re-equilibrated at the desired temperature. Several systems were expanded to a total of 288 lipids in order to improve the statistics for the analysis of alkane trajectories (for low alkane volume fractions), and to exclude interactions between different images of the same molecule[58] (for C30). For CHARMM36[32, 65] simulations, the DPPC force field was also taken from the open data by Miettinen and Ollila.[63] Water molecules were described by the CHARMM TIP3P model.[66, 67] For Slipids simulations, the DPPC parameters published as Supporting Information in the original Slipids publication were used.[34] In all simulations, alkane molecules were described by exactly the same force field parameters as the lipid acyl chains.
All simulations consisted of up to three runs (100 ns each), using a leap-frog integrator and 2 ps time steps. Most simulations were conducted in isothermic–isobaric (NPT) ensembles, except for two cases were the x and y box lengths were fixed (NPAT ensemble). Particle mesh Ewald was used for long-range electrostatics, and a fourth order LINCS correction for proton bond constraints. The detailed settings chosen for CHARMM36 and Slipids simulations are listed in Table 1. Details on the different algorithms used can be found in ref. [68] and references therein. CHARMM36 settings were based on the output of the CHARMM-GUI membrane builder.[41, 42, 69] The main simulation results are made available open source in the zenodo repository. These systems are summarized in Table S1, Supporting Information and can be accessed using the following DOI: https://doi.org/10.5281/zenodo.7572990. Additional systems discussed in the article include: 1) Pure, liquid n-alkanes C10, C20, and C30 (200 molecules per box), simulated at 298 K (C10), 323 K (C10 and C20), and 343 K (C30). Here, for both CHARMM36 and Slipids, the modified Berendsen thermostat (“v-rescale”) with a time constant τt=0.1 ps, and the Parrinello–Rahman barostat with a time constant τp=1.0 ps were used. 2) 10 vol% C20 in an NPAT ensemble, at two different areas-per-phospholipid (0.642 and 0.682 nm2, T = 60 °C, nw = 18), simulated using both CHARMM36 and Slipids. 3) 10 vol% C20 and pure DPPC at T = 60 °C, nw = 18, using CHARMM36 with the acyl chain charges of both alkane and lipid reduced by half. 4) A range of n-alkane simulations using identical conditions for CHARMM36 and Slipids (i.e., using the Slipids settings in Table 1 for CHARMM36 also). The investigated alkane concentrations were 0–30 vol% for C10, C20, and C30, at T = 70 °C and nw = 7/19. This data is not included in the zenodo repository but can be made available upon request.
Analysis of MD Simulations
The first 100 ns of each simulation were excluded from the analysis, and only 10 ps time steps were considered. The simulated trajectories were processed using Python's MDTraj library.[70]
Experimental Data
The experimental data from 2H and 1H-13C NMR measurements presented for comparison and assessment of the quality of the MD simulations was reused from a previous publication by our group.[29] Details on sample preparation and experimental setup can be found therein.
Acknowledgements
This research study was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG, project number 189853844, TRR 102). The authors acknowledge Wolfgang Paul, Jörg Kressler, and Markus Miettinen for useful discussions.
Open access funding enabled and organized by Projekt DEAL.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.