The impact of postsynaptic density 95 blocking peptide (Tat-NR2B9c) and an iNOS inhibitor (1400W) on proteomic profile of the hippocampus in C57BL/6J mouse model of kainate-induced epileptogenesis
Abstract
Antiepileptogenic agents that prevent the development of epilepsy following a brain insult remain the holy grail of epilepsy therapeutics. We have employed a label-free proteomic approach that allows quantification of large numbers of brain-expressed proteins in a single analysis in the mouse (male C57BL/6J) kainate (KA) model of epileptogenesis. In addition, we have incorporated two putative antiepileptogenic drugs, postsynaptic density protein-95 blocking peptide (PSD95BP or Tat-NR2B9c) and a highly selective inducible nitric oxide synthase inhibitor, 1400W, to give an insight into how such agents might ameliorate epileptogenesis. The test drugs were administered after the induction of status epilepticus (SE) and the animals were euthanized at 7 days, their hippocampi removed, and subjected to LC-MS/MS analysis. A total of 2,579 proteins were identified; their normalized abundance was compared between treatment groups using ANOVA, with correction for multiple testing by false discovery rate. Significantly altered proteins were subjected to gene ontology and KEGG pathway enrichment analyses. KA-induced SE was most robustly associated with an alteration in the abundance of proteins involved in neuroinflammation, including heat shock protein beta-1 (HSP27), glial fibrillary acidic protein, and CD44 antigen. Treatment with PSD95BP or 1400W moderated the abundance of several of these proteins plus that of secretogranin and Src substrate cortactin. Pathway analysis identified the glutamatergic synapse as a key target for both drugs. Our observations require validation in a larger-scale investigation, with candidate proteins explored in more detail. Nevertheless, this study has identified several mechanisms by which epilepsy might develop and several targets for novel drug development.
Open Practices
 This article has been awarded Open Data. All materials and data are publicly accessible as supporting information. Learn more about the Open Practices badges from the Center for Open Science: https://osf.io/tvyxz/wiki.
This article has been awarded Open Data. All materials and data are publicly accessible as supporting information. Learn more about the Open Practices badges from the Center for Open Science: https://osf.io/tvyxz/wiki.
Significance
Understanding the mechanisms that underlie the development of epilepsy has the potential to identify targets for antiepileptogenic agents that might prevent, delay or reverse the onset of spontaneous seizures. We have used the mouse kainate model of epileptogenesis and label-free proteomics to explore the effects of two mechanistically distinct and putative antiepileptogenic drugs. Our data support the contribution of neuroinflammation to epileptogenesis and its amelioration by drugs that target the glutamatergic synapse. Our findings offer unbiased insights into the events that occur during epileptogenesis and act as an important proof of concept for future, larger scale studies.
1 INTRODUCTION
Epileptogenesis is the process of developing epilepsy in response to altered gene expression or exposure to neurotoxins or head trauma (Pitkänen, Lukasiuk, Dudek, & Staley, 2015; Russo & Citraro, 2018). Epileptogenesis in humans and animal models have common features such as neuroinflammation, neurodegeneration, synaptic reorganization, and cognitive impairment (Beach, Woodhurst, Macdonald, & Jones, 1995; Koepp et al., 2017; Koh, 2018; Leal et al., 2017; Papageorgiou et al., 2018; Schmeiser, Zentner, Prinz, Brandt, & Freiman, 2017; Walker et al., 2017). We have previously demonstrated some of these in both rat and mouse models of kainate (KA)-induced temporal lobe epilepsy (TLE) (Puttachary, Sharma, Thippeswamy, & Thippeswamy, 2016; Puttachary, Sharma, Verma, et al., 2016).
In experimental models, chemoconvulsants such as the glutamate analog KA or the parasympathomimetic pilocarpine are commonly used to induce status epilepticus (SE). These models of TLE are well characterized, especially in rats with respect to gliosis, neurodegeneration, and spontaneous recurrent seizures (SRS). KA-induced SE in C57BL/6J (C57) mice triggers epileptogenesis but is rarely used as a TLE model for chronic studies due to infrequent SRS and insufficient evidence for all classical features of epileptogenesis. In our studies, continuous video-EEG recording for 4 months post-SE in C57 mice revealed a high frequency of convulsive SRS during the first 4–6 weeks post-SE, with electrographic non-convulsive seizures (NCS) persisting throughout (Puttachary et al., 2015). Immunohistochemistry (IHC) at 7, 14 and 28 days post-SE revealed significant gliosis, neurogenesis, and neurodegeneration in the hippocampus. In the current study, we chose 7 days post-SE, the maximum brain injury time point in C57 mice (Puttachary, Sharma, Thippeswamy, et al., 2016), to identify changes in protein expression in the hippocampus during epileptogenesis. We have used a label-free proteomics approach for this purpose. We have simultaneously investigated the effects of two mechanistically distinct putative antiepileptogenic agents; a highly selective inducible nitric oxide (NO) synthase (iNOS) inhibitor, 1400W, to target glial-derived NO and a postsynaptic density protein 95 (PSD95) blocking peptide (PSD95BP, also known as Tat-NR2B9c) to target neuronal hyperexcitability.
Our previous studies, and those of others, have shown that iNOS expression in hippocampus increases by 24–72 hr post-SE and persists for a long period (Cosgrave et al., 2008; Putra et al., 2019; Sharma et al., 2017; Vezzani, Aronica, Mazarati, & Pittman, 2013). Therefore, we hypothesized that iNOS is a potential promoter of epileptogenesis. Our studies in rats have also demonstrated that 1400W, when administered soon after KA-induced SE, significantly reduced neuroinflammation and epileptogenesis and also reduced neuropathic pain following nerve injury (Puttachary, Sharma, Verma, et al., 2016; Staunton, Barrett-Jolley, Djouhri, & Thippeswamy, 2018), suggesting its role in dampening neuronal hyperexcitability. In a recent organophosphate (OP) toxicity study, we showed that 1400W could suppress OP-induced upregulation of iNOS, 3-nitrotyrosine, gliosis, pro-inflammatory cytokines/chemokines, neurodegeneration, and SRS (Putra et al., 2019). While iNOS is predominantly expressed in glial cells in the CNS, PSD95 is localized to the postsynaptic neuronal cell membrane. PSD95 is a scaffolding protein and a potential target for postsynaptic modification (Swann et al., 2007; Wyneken et al., 2001). It is implicated in synaptic maturation and strengthening, and in plasticity, through its modulation of ionotropic glutamate receptors. Uncoupling of PSD95 from the NR2B subunit of the NMDA receptor disengages the intracellular cascade, affords neuroprotection in both pilocarpine-induced SE (Dykstra, Ratnam, & Gurd, 2009) and cerebral ischemia models (Aarts et al., 2002; Sun et al., 2008), and facilitates the isolation of excitotoxicity/neurodegeneration from epileptogenesis.
In this study, we investigated the effects of KA-induced SE and of 1400W and PSD95BP, when given alone and subsequent to SE, on the hippocampal proteomic profile in C57 mice at 7 days post-SE. Proteins whose expression was significantly altered in comparisons between treatment groups are reported and their potential contributions to epileptogenesis discussed.
2 MATERIALS AND METHODS
2.1 Animals
Adult male C57BL/6J mice (25–30 g) were purchased from Charles River Laboratories, Margate, UK. All animals were housed under standard conditions in the Biomedical Services Unit, University of Liverpool, with 12 hr light/dark cycle and access to food and water ad libitum. Animals were randomized and habituated in their housing environment in groups of five per cage for at least 7 days before any experimental procedures were performed. There were six treatment groups (n = 4 mice per group) as outlined in Figure 1. Full details of drug formulations and administration are provided in Sections 2.2 and 2.3. All in vivo experimental procedures were carried out in accordance with the Animals (Scientific Procedures) Act, UK (1986). Where appropriate, experiments were designed and have been reported in accordance with the principles of the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010).
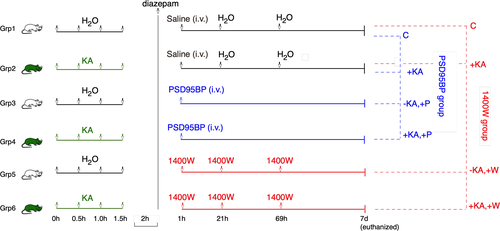
2.2 Induction of status epilepticus
Prior to the induction of SE, animals were weighed and then randomly assigned to individual cages, where food and water was available ad libitum. Animals were single-housed from the initiation of SE until the end of the experiment. The study employed behavioral indicators of seizure severity which were scored using a modified Racine scale (Racine, 2002) as follows: Stage 1, animals displayed mouth and facial twitching; Stage 2, animals displayed head nodding with myoclonic twitching and tremor; Stage 3, animals displayed forelimb clonus with a lordotic posture and Straub tail; Stage 4, animals reared with concomitant forelimb clonus; Stage 5, animals experienced a generalized clonic convulsion with wild running or jumping (Beamer, Otahal, Sills, & Thippeswamy, 2012; Hellier, Patrylo, Buckmaster, & Dudek, 1998; Macias et al., 2013; Puttachary et al., 2015; Schauwecker, 2010).
KA (Abcam, Cambridge, UK) was prepared fresh, as required, at a concentration of 2.5 mg/ml in sterile distilled water and sonicated for approximately 15 min until fully dissolved. Mice assigned to initial treatment with KA (i.e., Groups 2, 4, and 6) received 5 mg/kg KA (i.p. in 50–60 μl) every 30 min until the onset of the first Stage 5 seizure, at which point KA administration was discontinued. The vast majority of the mice required four injections of KA and only two mice required five injections to reach stage 5 seizures. The two mice that received five injections did not show any convulsive seizures prior to the fifth dose. We have previously shown that this method of SE induction, with repeated low dose (RLD) of KA, produces convulsive seizures lasting for >30 min with minimal mortality during the 2 hr of SE (Puttachary et al., 2015; Sharma et al., 2018; Tse et al., 2014). The control animals (i.e., those in Groups 1, 3, and 5) received four injections (i.p.) of 50–60 μl distilled water separated by 30 min each to simulate KA administration. Two hours after the onset of the first Stage 5 seizure (or after the fourth injection of distilled water in control animals), all mice received 10 mg/kg diazepam in 50–60 μl (i.p.) to standardize the duration of KA-induced SE. Diazepam was obtained from Hameln Pharmaceuticals, Gloucester, UK by the Named Veterinary Surgeon, Biomedical Services Unit, University of Liverpool. We and others have previously shown that the use of diazepam at this dose and time point (>45 min of continuous SE) does not interfere with the subsequent onset of epileptogenesis or neuropathology (Aroniadou-Anderjaska, Fritsch, Qashu, & Braga, 2008; Beamer et al., 2012; Ben-Ari, Tremblay, Ottersen, & Meldrum, 1980; Puttachary, Sharma, Thippeswamy, et al., 2016; Puttachary et al., 2015). After diazepam administration, mice were returned to their individual cages prior to further intervention.
2.3 Drug treatments
Mice in Groups 1 and 2 (n = 4 per group) served as treatment controls for drug interventions (Figure 1). Each animal received a single administration of 75 μl sterile saline (i.v.), the vehicle for PSD95BP, into the tail vein at 1 hr after diazepam treatment and two further injections (i.p.) of 125–150 μl sterile distilled water, the vehicle for 1400W, at 21 and 69 hr after diazepam.
PSD95BP (Tat-NR2B9c) was kindly provided by Professor Tymianski at the University of Toronto. It comprises nine C-terminal residues [KLSSIESDV] of the NR2B subunit of the NMDA subtype of glutamate receptor (NMDAR) conjugated to the cell membrane protein transduction domain of a human immunodeficiency virus type 1 Tat protein, which facilitates the blood–brain barrier permeability (Aarts et al., 2002; Cook, Teves, & Tymianski, 2012; Cui et al., 2007; Forder & Tymianski, 2009; Schwarze, Ho, Vocero-Akbani, & Dowdy, 1999; Sun et al., 2008). In this study, PSD95BP was prepared in sterile saline solution (0.9% NaCl) and administered as a single bolus dose of 7.6 mg/kg in 75 μl (i.v.) via the tail vein to a total of eight mice (i.e., all animals in Groups 3 and 4) at 1 hr after diazepam administration (Figure 1). In previous studies using rodent models, this dose has been shown to interact with the PDZ domain of PSD95 and neuronal nitric oxide synthase (nNOS), and to disengage the PSD95-nNOS complex from NMDAR without affecting NMDAR-mediated currents (Cui et al., 2007).
About 1400W dihydrochloride (Tocris Bioscience, Bristol, UK) was prepared at a concentration of 4 mg/ml in sterile distilled water and administered to a total of eight mice (i.e., all animals in Groups 5 and 6) at a dose of 20 mg/kg (i.p.) at 1 hr after diazepam treatment. This was followed by two further administrations, again at a dose of 20 mg/kg in 125–150 μl (i.p.), at 21 and 69 hr thereafter (i.e., at 24 and 72 hr after the onset of the first Stage 5 seizure; Figure 1). The dose and treatment schedule were chosen on the basis of our previous work on NO (and that of others) and modified for use in mice. It has been shown that KA-induced pro-inflammatory cytokines and NO occurs in phases after SE (Ravizza et al., 2005; Ryan et al., 2014). We had also observed a reduction in iNOS expression in glial cells and gliosis at 72 hr in the animals treated with NOS inhibitors, after induction of SE, in the rat and C57 mouse models (Beamer et al., 2012; Cosgrave et al., 2008).
2.4 Tissue preparation
Seven days after induction of SE, all mice were euthanized with an overdose of sodium pentobarbitone (Pentoject®; 60 mg/kg, i.p.) and death confirmed by exsanguination. Sodium pentobarbitone was obtained from Animalcare Limited (York, UK) by the Named Veterinary Surgeon, Biomedical Services Unit, University of Liverpool. Brains were rapidly removed postmortem and the hippocampi dissected on ice and placed in chilled cryovials prior to being snap-frozen in liquid nitrogen. All tissue samples were coded and stored at −80°C until required for proteomic analysis.
2.5 Sample preparation for proteomic analysis
All chemicals and solvents (HPLC grade) were obtained from Sigma Aldrich (Gillingham, UK) unless otherwise stated. Hippocampi were removed from the storage, thawed on ice, and homogenized directly in the storage cryovial in 0.5 ml of ice-cold 25 mM of NH4HCO3 for 30 s using a Tissue Ruptor (Qiagen, UK). A 15 μl aliquot of each homogenate (equal to 50–100 μg protein) was added to a low-bind tube and diluted to 170 μl using 0.06% (w/v) Rapigest SF (Waters MS Technologies, Manchester, UK) in 25 mM of NH4HCO3. Samples were stored at −20°C until all homogenizations were complete. Samples were then thawed, heated to 80°C for 10 min, reduced by the addition of 10 l of 60 mM dithiothreitol (Melford Laboratories, Ipswich, UK) in 25 mM of NH4HCO3, and incubated at 60°C for 10 min. Samples were subsequently alkylated by the addition of 10 μl of 180 mM of iodoacetamide and incubated at room temperature in the dark for 30 min. Thereafter, 10 μl of 0.2 μg/μl of trypsin (in 50 mM of acetic acid) was added and samples incubated overnight at 37°C with intermittent shaking. Following overnight incubation, the protein content of each sample was assayed using a modification of the Bradford assay (Bradford, 1976). Rapigest SF was then removed by the addition of 2 µl of 99% (v/v) trifluoroacetic acid and the samples incubated at 37°C for 45 min. Samples were centrifuged at 17,200 × g for 30 min and supernatants transferred to fresh 0.5 ml low-bind tubes. The centrifugation step was repeated and 10 μl of the final supernatant was then transferred to a total recovery vial (Waters, UK) for proteomic analysis by liquid chromatography-mass spectrometry (LC-MS).
2.6 Proteomic analysis
All peptide separations were carried out using an Ultimate 3000 Nanosystem (Dionex/Thermo Fisher Scientific, UK). For each analysis, 1 μg (protein equivalent) of sample was loaded onto a trapping column (Acclaim PepMap 100, 2 cm × 75 μm inner diameter, C18, 3 μm, 100 Å) at 5 μl/min with an aqueous solution containing 0.1% (v/v) trifluoroacetic acid and 2% (v/v) acetonitrile. After 3 min, the trap column was set in-line with an analytical column (Easy-Spray PepMap® RSLC 50 cm × 75 μm inner diameter, C18, 2 μm, 100 Å) (Dionex, UK). Peptide elution was performed by applying a mixture of solvents A and B. Solvent A was HPLC grade water with 0.1% (v/v) formic acid, and solvent B was 80% acetonitrile (v/v) with 0.1% (v/v) formic acid. Separations were performed by applying a linear gradient of 3.8%–40% solvent B over 90 min at 300 nl/min followed by a washing step (5 min at 99% solvent B) and an equilibration step (15 min at 3.8% solvent B).
The Q Exactive instrument was operated in data-dependent positive (ESI+) mode to automatically switch between full scan MS and MS/MS acquisition. Survey full scan MS spectra (m/z 300–2000) were acquired in the Orbitrap with 70,000 resolution (m/z 200) after accumulation of ions to 1 × 106 target value based on predictive automatic gain control (AGC) values from the previous full scan. Dynamic exclusion was set to 20 s. The 10 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the octopole collision cell by higher energy collisional dissociation (HCD) with a fixed injection time of 50 ms and resolution of 17,500. Typical mass spectrometric conditions were as follows: spray voltage 2.1 kV, no sheath or auxillary gas flow; heated capillary temperature 250°C; normalized HCD collision energy 30%. The MS/MS ion selection threshold was set to 2 × 104 counts and a 2 m/z isolation width was set.
2.7 Data analysis
Raw data files were uploaded into Progenesis LC-MS version 3.0 (Nonlinear Dynamics Ltd, UK) for automatic alignment and peak picking. Data were filtered with peptide charges of +1 and >7 removed. The resulting peak list was submitted to the Mascot search engine (version 2.6; Matrix Science, UK) and searched against a reviewed Mouse UniProt protein database (date: 15/02/17; containing 16,843 entries) with carbamidomethyl cysteine as a fixed modification and oxidation of methionine as a variable modification. Peptide mass tolerance was 10 ppm and fragment mass tolerance was 0.01 Da. A total of 2,579 protein families were returned (raw data available by request to corresponding authors).
Data were analyzed independently by drug for both PSD95BP (Group 1—VEH, Group 2—KA, Group 3—VEH + PSD, Group 4—KA + PSD) and 1400W (Group 1—VEH, Group 2—KA, Group 5—VEH + 1400W, and Group 6—KA + 1400W), with statistical comparison between individual treatment groups in each analysis carried out using ANOVA within the Progenesis software. All proteins showing a statistically significant (p < 0.05) change in normalized abundance between treatment groups were identified and reported on the basis of the number of peptides (including unique peptides) contributing to the analysis, p-value (uncorrected), false discovery rate (FDR) adjusted q-value, maximum fold change (Δ), and direction of effect. Lists of differentially expressed proteins were subject to Gene Ontology (GO) and KEGG pathway enrichment analysis using the R package “clusterProfiler” (v3.10.1; Yu, Wang, Han, & He, 2012) against the complete mouse annotation. KEGG pathway diagrams annotated with abundance data were generated using the KEGG pathview package (v1.22.30; Luo & Brouwer, 2013).
3 RESULTS
3.1 Outcome of KA administration
All KA-exposed animals experienced convulsive seizures (≥stage 3) for >30 min during 2 hr of SE duration recorded from the time of onset of the first Stage 5 seizure to the termination with diazepam treatment. Seizure severity during SE was monitored visually in real-time and quantified as described previously (Beamer et al., 2012; Tse et al., 2014). There were no significant differences in SE severity between animals that were allocated to Group 2, Group 4, and Group 6 (Figure 2). After termination of SE with diazepam, KA-treated animals and their matched controls were randomly allocated to treatment with either PSD95BP, 1400W or vehicle alone (as detailed in Figure 1). All interventions were applied after termination of behavioral SE to ensure that they did not modify the initial insult (i.e., severity of SE).
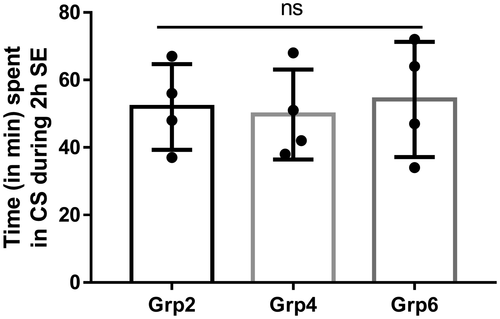
All mice survived in this study due to the RLD method of SE induction by KA. However, as expected the animals lost bodyweight during the first 48–72 hr post-SE. Sterile dextrose saline supplement (1 ml, s.c.) twice daily for the first 3 days helped to regain the bodyweight. In the 7 days between KA-induced SE and harvesting of tissue, animals were inspected visually on a twice-daily basis as per standard husbandry practice. There were no overt signs of seizure activity in any of the animals during visual inspection, and no notable behavioral sequelae of KA and/or drug treatment. We were not able to incorporate continuous video-EEG monitoring in the current study due to the limited capacity (at that time) for contemporaneous recording. However, we have previously reported increased epileptiform spiking, numerous electrographic NCS, and occasional SRS during the first 7 days post-SE using the RLD method of KA administration to C57 mice (Puttachary et al., 2015).
3.2 Effect of PSD95BP on the hippocampal proteome following KA-induced SE
The effects of PSD95BP were investigated by the combined analysis of proteomic data from four groups of animals; Group 1 (VEH), Group 2 (KA), Group 3 (VEH + PSD), and Group 4 (KA + PSD). Of 2,579 proteins detected in this study (see supplementary boxplots file), 175 individual proteins showed a significant change in abundance (p < 0.05) as a result of intervention with PSD95BP following KA-induced SE (Supplementary Table S1) but only 11 of those changes in abundance survived correction for multiple testing (q < 0.05; Table 1).
| Accession | Peptides | Mascot Score | % Seq Coverage | p-value | q-value | Fold change | High group | Low group | Description |
|---|---|---|---|---|---|---|---|---|---|
| P14106 | 4 | 706 | 19.4 | 4.26E−06 | 0.0104 | 1.6 | KA | VEH | C1QB_MOUSE Complement C1q subcomponent subunit B |
| Q9QUR6 | 8 | 1,230 | 17.2 | 1.40E−05 | 0.0132 | 1.534 | KA | VEH + PSD | PPCE_MOUSE Prolyl endopeptidase |
| P15379 | 1 | 451 | 1.5 | 2.04E−05 | 0.0132 | 4.941 | KA | VEH + PSD | CD44_MOUSE CD44 antigen |
| P03995 | 36 | 22,690 | 70.9 | 2.54E−05 | 0.0132 | 2.611 | KA | VEH | GFAP_MOUSE Glial fibrillary acidic protein |
| P14602 | 2 | 43 | 12.4 | 2.69E−05 | 0.0132 | 4.076 | KA | VEH | HSPB1_MOUSE Heat shock protein beta−1 |
| P26041 | 8 | 896 | 16.8 | 5.65E−05 | 0.0231 | 1.724 | KA | VEH | MOES_MOUSE Moesin |
| P21460 | 6 | 1953 | 43.6 | 0.000102 | 0.0351 | 1.381 | KA | VEH + PSD | CYTC_MOUSE Cystatin-C |
| P16045 | 4 | 609 | 33.3 | 0.000125 | 0.0351 | 1.957 | KA | VEH | LEG1_MOUSE Galectin−1 |
| P20152 | 28 | 11,291 | 59.4 | 0.000139 | 0.0351 | 1.727 | KA | VEH | VIME_MOUSE Vimentin |
| Q03517 | 10 | 975 | 24.6 | 0.000143 | 0.0351 | 2.059 | KA + PSD | VEH | SCG2_MOUSE Secretogranin−2 |
| Q99L04 | 7 | 1,495 | 32.9 | 0.000205 | 0.0456 | 1.922 | KA | VEH | DHRS1_MOUSE Dehydrogenase/reductase SDR family member 1 |
Seven of those 11 proteins were derived from comparison of data from Group 1 (VEH) and Group 2 (KA) and were accordingly considered reflective of the effects of KA alone (Table 1). In all seven cases, protein abundance was significantly higher in Group 2 (KA) than in Group 1 (VEH), suggesting increased expression as a result of KA-induced SE at 7 days prior to harvesting of tissue. Two of seven proteins whose abundance was significantly elevated by KA had a fold-change (Δ) that exceeded 2.00; heat shock protein beta-1 (Hspb1/Hsp27; Δ = 4.08, q = 0.0132; Figure 3) and glial fibrillary acidic protein (Gfap/GFAP; Δ = 2.61, q = 0.0132; Figure 3).
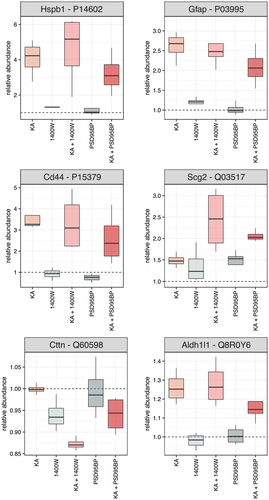
The remaining four proteins that showed a statistically significant alteration in abundance were related to PSD95BP treatment (Table 1). In three of four, abundance was highest in Group 2 (KA) and lowest in Group 3 (VEH + PSD), suggesting that treatment with PSD95BP alone reduced the expression of a protein that was otherwise elevated by KA-induced SE. Of those three, the greatest change in abundance was seen with CD44 antigen (Δ = 4.94, q = 0.0132; Figure 3). There was also a statistically significant change in the abundance of secretogranin (Δ = 2.06, q = 0.0351; Figure 3), with lowest abundance in Group 1 (VEH) and highest in Group 4 (KA + PSD).
3.3 Effect of 1400W on the hippocampal proteome following KA-induced SE
The effects of 1400W were investigated by the combined analysis of proteomic data from four groups of animals; Group 1 (VEH), Group 2 (KA), Group 5 (VEH + 1400W), and Group 6 (KA + 1400W). Of 2,579 proteins detected in this study (see supplementary boxplots file), 168 individual proteins showed a significant change in abundance (p < 0.05) as a result of intervention with 1400W following KA-induced SE (Supplementary Table S2) but only 12 of those changes in abundance survived correction for multiple testing (q < 0.05; Table 2).
| Accession | Unique | Mascot Score | % Seq Coverage | p-value | q-value | Fold change | High group | Low group | Description |
|---|---|---|---|---|---|---|---|---|---|
| P14106 | 3 | 706 | 19.4 | 7.18E−07 | 0.0013 | 1.6 | KA | VEH | C1QB_MOUSE Complement C1q subcomponent subunit B |
| P21460 | 6 | 1953 | 43.6 | 1.09E−06 | 0.0013 | 1.452 | KA | VEH + 1400W | CYTC_MOUSE Cystatin-C |
| P26041 | 5 | 896 | 16.8 | 1.48E−06 | 0.0013 | 1.906 | KA + 1400W | VEH | MOES_MOUSE Moesin |
| P03995 | 28 | 22,690 | 70.9 | 9.56E−06 | 0.0062 | 2.611 | KA | VEH | GFAP_MOUSE Glial fibrillary acidic protein |
| P16045 | 4 | 609 | 33.3 | 1.32E−05 | 0.0062 | 1.957 | KA | VEH | LEG1_MOUSE Galectin−1 |
| P15379 | 1 | 451 | 1.5 | 1.45E−05 | 0.0062 | 3.917 | KA | VEH + 1400W | CD44_MOUSE CD44 antigen |
| Q60598 | 13 | 3,124 | 28.8 | 2.39E−05 | 0.0088 | 1.146 | VEH | KA + 1400W | SRC8_MOUSE Src substrate cortactin |
| P14602 | 2 | 43 | 12.4 | 3.41E−05 | 0.011 | 4.605 | KA + 1400W | VEH | HSPB1_MOUSE Heat shock protein beta−1 |
| Q8R0Y6 | 23 | 5,970 | 38 | 4.84E−05 | 0.0139 | 1.305 | KA + 1400W | VEH + 1400W | AL1L1_MOUSE Cytosolic 10-formyltetrahydrofolate dehydrogenase |
| Q02105 | 3 | 395 | 16.3 | 6.06E−05 | 0.0156 | 1.727 | KA | VEH | C1QC_MOUSE Complement C1q subcomponent subunit C |
| Q99L04 | 7 | 1,495 | 32.9 | 0.000102 | 0.0239 | 1.922 | KA | VEH | DHRS1_MOUSE Dehydrogenase/reductase SDR family member 1 |
| P20152 | 24 | 11,291 | 59.4 | 0.000129 | 0.0276 | 1.838 | KA + 1400W | VEH | VIME_MOUSE Vimentin |
Five of those 12 proteins were derived from comparison of data from Group 1 (VEH) and Group 2 (KA) and were again considered reflective of the effects of KA alone (Table 2). In all five cases, protein abundance was significantly higher in Group 2 (KA) than in Group 1 (VEH), suggesting increased expression as a result of KA-induced SE at 7 days prior to harvesting of tissue. Just one of five proteins whose abundance was significantly elevated by KA had a fold-change that exceeded 2.00; GFAP (Δ = 2.61, q = 0.0062; Figure 3).
The remaining seven proteins that showed a statistically significant alteration in abundance were related to 1400W treatment (Table 2). Three of those showed highest abundance in Group 6 (KA + 1400W) and lowest in Group 1 (VEH), suggesting a combined action of KA-induced SE and 1400W on protein expression, the most notable of which was a 4.61-fold increase in abundance of Hsp27 (q = 0.011; Figure 3). Two proteins showed highest abundance in KA-treated mice (Group 2) and lowest in those treated with 1400W alone (Group 4), suggesting a suppressive action of the drug on the expression of a protein that was otherwise elevated by KA-induced SE; this was exemplified by CD44 antigen (Δ = 3.92, q = 0.0062; Figure 3). Finally, there were two proteins that showed modest (<1.5) but nonetheless significant fold-changes in abundance; Src substrate cortactin (q = 0.0088; Figure 3), which was highest in Group 1 (VEH) and lowest in Group 6 (KA + 1400W), and cytosolic 10-formyltetrahydrofolate dehydrogenase (Aldh1L1/AL1L1, q = 0.0139; Figure 3), which was highest in Group 6 (KA + 1400W) and lowest in Group 4 (VEH + 1400W).
3.4 Pathway analysis
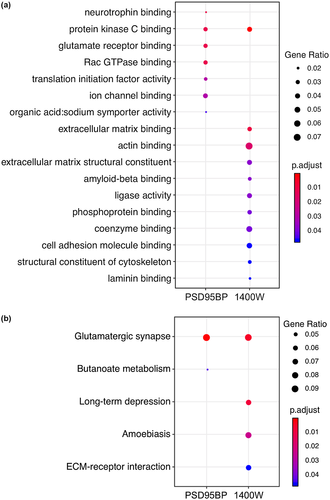
Individual proteins whose abundance was significantly (ANOVA p < 0.05) altered in the PSD95BP analysis (Supplementary Table S1) and 1400W analysis (Supplementary Table S2) were subjected to gene ontology (GO) and KEGG pathway enrichment analyses against the complete mouse annotation. The most enriched GO terms, based on a hypergeometric distribution, among differentially abundant proteins in the PSD95BP data set included those related to “neurotrophin binding,” “glutamate receptor binding,” “ion channel binding,” “translation initiation factor activity,” “Rac GTPase binding,” and “protein kinase C binding” (Figure 4a). KEGG pathway analysis on the PSD95BP data set revealed “glutamatergic synapse” and “butanoate metabolism” to be the most enriched pathways (Figure 4b). Enriched GO terms among differentially abundant proteins in the 1400W data set included those related to “protein kinase C binding,” “actin/cytoskeletal binding,” “extracellular matrix,” “amyloid-beta binding,” “ligase activity,” “phosphoprotein binding,” “coenzyme binding,” “cell adhesion molecule binding,” and “laminin binding” (Figure 4a). KEGG pathway analysis on the 1400W data set again revealed terms relating to “glutamatergic synapse” as the most prominent, but also terms linked to “long-term depression” and “extra-cellular matrix interactions” (Figure 4b). Full lists of significantly enriched GO terms and KEGG pathways and their associated proteins are provided as Supplementary Tables S3 and S4. Visualizations of “glutamatergic synapse” pathways for PSD95BP and 1400W generated in the KEGG pathview package and annotated with corresponding protein abundance data are additionally provided in Supplementary Figure S1 (PSD95BP) and S2 (1400W).
3.5 Box-and-Whisker plots for all proteins
A comprehensive supplementary data for all 2,579 proteins are included as Supplementary Figure S3.
4 DISCUSSION
Previous studies have provided important insights into the underlying cellular and molecular processes of epileptogenesis, highlighting neuroinflammation, the innate immune response, microglial activation, synaptic reorganization, and oxidative stress as key mechanisms in this regard (Bitsika et al., 2016; Keck et al., 2017, 2018; Li et al., 2010; Liu et al., 2008; Marques-Carneiro et al., 2017; Walker et al., 2016). Many of these observations are unsurprising, given the burgeoning knowledge base surrounding the mechanisms and pathways believed to contribute to the development of epilepsy (Aronica et al., 2017; Becker, 2018; Godale & Danzer, 2018; Lukawski et al., 2018; Pitkänen et al., 2015). Our experimental design is novel with respect to the choice of two putative anti-epileptogenic agents, that is, PSD95BP (Tat-NR2B9c) that targets neurons and 1400W that targets the glial source of NO. This approach not only allowed an assessment of their molecular effects in normal brain but also an opportunity to explore their effects during the epileptogenic period.
The KA mouse model of epileptogenesis is widely employed in experimental epilepsy research. We have used a repeated, low dose approach to the administration of KA that successfully resulted in sustained SE in all exposed animals, as described in our previous work (Puttachary et al., 2015; Tse et al., 2014). There were no significant differences in the severity of the initial SE in any of the relevant treatment groups, limiting the impact of insult heterogeneity on subsequent protein expression, and interventions (i.e., PSD95BP and 1400W) were administered at least 1 hr after the termination of SE to avoid the possibility of insult modification. Analysis of isolated hippocampal tissue at 7 days post-SE using a label-free proteomics approach detected a total of 2,579 unique proteins. Initial statistical analysis, stratified by drug treatment, identified approximately 170 proteins in each data set whose abundance was significantly altered (ANOVA p < 0.05). However, following appropriate correction for multiple testing, only a handful of proteins remained (Tables 1 and 2). Of those, several were common to both data sets and had lowest abundance in vehicle-treated animals (Group 1) and highest in the KA group (Group 2). They were accordingly considered to reflect the impact of KA-induced SE rather than intervention with either PSD95BP or 1400W. Those proteins included complement C1q subcomponent subunit B (C1qB), GFAP, and galectin-1, all of which have previously been implicated in epileptogenesis (Bischoff, Deogracias, Poirier, & Barde, 2012; Schartz, Wyatt-Johnson, Price, Colin, & Brewster, 2018).
There is very recent evidence suggesting that C1q activation in microglia is an important mediator of neuroinflammation and epileptogenesis (Schartz et al., 2018). C1q proteins expressed during epileptogenesis also serve as extracellular organizers, responsible for recruitment, and clustering of functional KA receptors on the postsynaptic membrane in hippocampal pyramidal neurons (Matsuda et al., 2016), suggesting a role in the perpetuation of SRS and the possibility that they might be viable targets for antiepileptogenic drug development. However, it should also be noted that C1q knockout mice develop epilepsy as a result of enhanced synaptic connectivity (Chu et al., 2010), implying that functional C1q proteins are required for effective synaptic pruning during development to establish normal connectivity (Ma, Ramachandran, Ford, Parada, & Prince, 2013). As with C1qB, an increase in the abundance of GFAP is consistent with the activation of astrocytes and microglial cells that is routinely observed following brain insults (Ghirnikar, Lee, & Eng, 1998). This finding lends further weight to the importance of glial cells in the early stages of epileptogenesis, by virtue of their role in neuroinflammation (Puttachary, Sharma, Verma, et al., 2016; Sharma et al., 2018; Terrone, Salamone, & Vezzani, 2017; Walker et al., 2017). It also serves to validate our experimental paradigm, as a lack of change in GFAP expression might have questioned the robustness of our model and/or the sensitivity of the data analysis. Less is known about the role of galectin-1 in epileptogenesis but there is some evidence that it can regulate neuronal cell death in the pilocarpine model of epilepsy (Bischoff et al., 2012).
The majority of the statistically significant changes in individual protein abundance observed in this study were believed to be driven by KA-induced SE, rather than by treatment with either PSD95BP or 1400W. This is perhaps not surprising, given the known neuropathological effects of KA exposure (Benkovic, O'Callaghan, & Miller, 2006; Puttachary, Sharma, Thippeswamy, et al., 2016). Nevertheless, analysis of the raw Progenesis output, as reported in Tables 1 and 2, did identify a number of instances where changes in protein abundance were associated with drug treatment groups in one or both data sets.
The two proteins with the greatest fold-changes in abundance were Hsp27 and CD44 antigen. The abundance of Hsp27 was highest in Group 6 animals (KA + 1400W) and lowest in Group 1 (VEH), perhaps suggestive of an endogenous protective effect against the KA insult that is somehow augmented in the presence of iNOS inhibition. Hsp27 is considered to be a protector of cells and sub-cellular organelles, including mitochondria, and its expression is known to be induced under various pathological conditions (Franklin, Krueger-Naug, Clarke, Arrigo, & Currie, 2005). Transgenic mice that overexpress Hsp27 are known to be resistant to KA-induced neurodegeneration suggesting its neuroprotective effects (Akbar et al., 2003). However, interestingly, it is also considered as a reliable marker of SE severity in the pilocarpine epilepsy model (Kirschstein et al., 2012), and is significantly up-regulated in resected neocortex from people with drug resistant epilepsy (Bidmon et al., 2004). CD44 antigen is cell surface glycoprotein that is traditionally considered to promote cell-cell interactions and cell migration and adhesion (Puré & Cuff, 2001). Recent evidence suggests that this role extends to maintenance of the structural and functional plasticity of dendritic spines (Roszkowska et al., 2016). The robust change in abundance of CD44 antigen, which was significantly elevated in Group 2 (KA) animals compared to drug treatment groups (Group 3, VEH + PSD; Group 5, VEH + 1400W), suggests either the involvement of the innate immune response (mediated by T-cell migration) or an ongoing synaptic re-organization in the aftermath of KA-induced SE, both of which are plausible. Interestingly, an upregulation of hippocampal CD44 antigen has been also reported at 2 and 10 days post-SE in a rat model of epilepsy (Keck et al., 2018).
Other proteins that showed statistically significant changes in abundance associated with drug treatment included moesin, vimentin, Src substrate cortactin, and secretogranin-2. Again, there is evidence to support a role for each of these proteins in epileptogenesis. Moesin has previously shown differential expression in sclerotic versus non-sclerotic human hippocampi from epilepsy patients (Lee et al., 2007) and upregulation in epilepsy-associated glioneuronal lesions (Majores et al., 2005). Likewise, vimentin, an intermediate filament protein that co-localizes with GFAP in reactive astrocytes (Sancho-Tello, Vallés, Montoliu, Renau-Piqueras, & Guerri, 1995), is elevated in seizures associated with angiocentric glioma (Ni et al., 2015) and is also considered to be a marker of the astrogliosis seen in tuberous sclerosis (Sosunov et al., 2008). A change in abundance of Src substrate cortactin is consistent with the involvement of Src kinases and neuroinflammation in epileptogenesis, which we have demonstrated previously (Sharma et al., 2018), while secretogranin-2 is up-regulated following experimental seizures, particularly with KA, in a distinctive cell-type and species specific manner (Kandlhofer et al., 2000; Mahata et al., 1992; Marti, Blasi, & Ferrer, 2002).
All of the changes observed in individual protein abundance (i.e., C1q, GFAP, Hsp27, CD44 antigen, etc.) in this study were related to the processes of neuroinflammation and the activation of glial cells and the innate immune response, which served to confirm much of what is currently understood about the events underlying epileptogenesis. Interestingly, however, pathway analysis instead identified glutamate receptor binding, protein kinase C binding, cytoskeletal aspects, and extracellular matrix interactions as the most significantly enriched GO terms and the glutamatergic synapse as the most enriched pathway. Accordingly, one might speculate that the individual protein changes reflected the dominant effect of KA-induced seizures, while the functional analysis, based on an unbiased, label-free proteomics analysis, was more sensitive to the pharmacology of the drugs employed in this study, both of which are known to target the glutamatergic synapse.
This study has afforded an opportunity to identify proteins whose abundance is changed following KA-induced SE, and also following treatment with both PSD95BP and 1400W. Part of the rationale for this study was also to explore how those drugs might interfere with, and perhaps protect against, the epileptogenic process initiated by KA. That aim was predicated on two important assumptions; firstly that KA exposure is epileptogenic in the animals employed in this study, and that PSD95BP and 1400W have antiepileptogenic properties. Our previous work supports the development of NCS and occasional SRS in the C57BL/6J mouse following KA-induced SE (Puttachary et al., 2015), but we did not specifically observe spontaneous seizures in the current 7 days post-SE study. Likewise, there is existing evidence that PSD95BP and 1400W have disease-modifying properties in epilepsy models (Dykstra et al., 2009; Puttachary, Sharma, Thippeswamy, et al., 2016; Putra et al., 2019) but, in the absence of overt epileptogenesis in the current study, it is not possible to prove that these drugs were antiepileptogenic on this occasion.
Few studies have explored the proteomic signature of epileptogenesis to date (Bitsika et al., 2016; Keck et al., 2017, 2018; Li et al., 2010; Liu et al., 2008; Marques-Carneiro et al., 2017; Walker et al., 2016), all of which have been relatively small scale investigations involving small numbers of animals. In this respect, the current study is no different. Nevertheless, we have highlighted several key proteins that are seemingly perturbed by the process of epileptogenesis. Some of those proteins are already well-recognized (i.e., GFAP and C1q), whereas others (i.e., Hsp27 and CD44 antigen) have not been widely reported previously in epilepsy models. Most are involved in neuroinflammation, which is widely believed to underlie the development of epilepsy, and the net effect of those changes in protein abundance appears to map to the glutamatergic synapse, which is the target of the experimental therapeutics used in this study. Our observations clearly require validation in a larger-scale investigation, with candidate proteins explored in more detail. This is only an exploratory study in males but more research is needed including in females.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
ACKNOWLEDGMENTS
The authors would like to thank the Biotechnology and Biological Sciences Research Council (UK) for funding this study, the Institute of Ageing and Chronic Disease, University of Liverpool for their support of a PhD studentship to KT, and the NIH (NINDS/CounterACT) funding support to TT (1R21NS099007-01A1).
CONFLICT OF INTEREST
There are no conflicts of interest.
AUTHOR CONTRIBUTIONS
TT conceived the idea, wrote the grant application (GJS as a Co-Investigator), secured the funding, recruited collaborators and reagents, obtained the project license from the UK Home Office, designed experiments in consultation with RJB, recruited KT and EB as PhD students, trained, supervised in vivo experiments. TT and GJS wrote/edited the manuscript. KT, EB, and DS conducted the experiments. KT, DS, DH, and GJS analyzed the data. RJB contributed to the design of the study and supervised proteomics experiments. RJB, DS, and DH wrote the proteomics methodology and drafted Figure 1. TT drafted the Graphical abstract. MT and MS kindly supplied PSD95BP for the project.