miR-34a/c induce caprine endometrial epithelial cell apoptosis by regulating circ-8073/CEP55 via the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways
Xiaorui Liu, Lei Zhang, and Lichun Yang contributed equally to this study.
Abstract
microRNAs (miRNAs) and circular RNAs (circRNAs) are important for endometrial receptivity establishment and embryo implantation in mammals. miR-34a and miR-34c are highly expressed in caprine receptive endometrium (RE). Herein, the functions and mechanisms of miR-34a/c in caprine endometrial epithelial cell (CEEC) apoptosis and RE establishment were investigated. miR-34a/c downregulated the expression level of centrosomal protein 55 (CEP55) and were sponged by circRNA8073 (circ-8073), thereby exhibiting a negative interaction in CEEC. miR-34a/c induced CEEC apoptosis by targeting circ-8073/CEP55 through the regulation of the RAS/RAF/MEK/ERK and phosphoitide 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathways. Positive and negative feedback loops and cross-talk were documented between the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways. miR-34a/c regulated the levels of RE marker genes, including forkhead box M1, vascular endothelial growth factor, and osteopontin (OPN). These results suggest that miR-34a/c not only induce CEEC apoptosis by binding to circ-8073 and CEP55 via the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways, but may also regulate RE establishment in dairy goats.
1 INTRODUCTION
Embryo implantation, a critical step in the establishment of pregnancy, requires a receptive endometrium (RE) in mammals (Achache & Revel, 2006; Schulte, Tsai, & Moley, 2015). Formation of RE is a complicated and dynamic process during which the endometrium undergoes morphological and physiological changes (S. Zhang et al., 2013). Apoptosis of caprine endometrial epithelial cells (CEECs) is an indispensable physiological event enabling the embryo to breach the epithelial barrier and implant within the luminal epithelium of the uterus (X. Wang, Wu, & Demayo, 2017). Understanding the molecular mechanisms regulating CEEC apoptosis during embryo implantation is essential to elucidate RE establishment in dairy goats.
The first microRNA (miRNA) profile of RE biology during caprine embryo implantation suggested that a subset of miRNAs might play important roles in the establishment of caprine RE (Y. Song et al., 2015). Our previous studies have demonstrated that miRNAs can regulate caprine endometrial cell proliferation or apoptosis, and promote RE establishment (An et al., 2017; L. Zhang, Liu, Liu, Zhou, et al., 2017). For instance, we discovered that miR-26a regulates caprine endometrial cell apoptosis or proliferation (Lei et al., 2017), and that miR-449a plays a vital role in CEEC and the establishment of caprine RE (An et al., 2017; Liu et al., 2018). Additionally, our previous sequencing results showed that miR-34a (miR-34a-5p) and miR-34c (miR-34c-5p) were 2.55- and 3.04-fold higher, respectively, in RE than in pre-receptive endometrium (PE; Y. Song et al., 2015). It must be noted that miR-449a, miR-34a, and miR-34c are members of the miR-34/449 superfamily (Mercey et al., 2017) with strong sequence homology, and their shared seed sequence may be indicative of shared targets (Ebner & Selbach, 2014; R. Song et al., 2014). However, the functions and mechanisms of miR-34a/c in CEEC and RE establishment remain largely unknown.
Circular RNAs (circRNAs) can act as posttranscriptional regulators by sponging miRNAs (Memczak et al., 2013), and some circRNAs may have important roles in CEEC and RE establishment (L. Zhang, Liu, Che, et al., 2018; L. Zhang, Liu, Ma, et al., 2018). For example, Circ-3175 functions as a competing endogenous RNA that suppresses miR-182, thereby protecting testin and inhibiting CEEC apoptosis (L. Zhang, Liu, Ma, et al., 2018). Additionally, circ-8073 was shown to inhibit CEEC apoptosis by sponging miR-449a to upregulate CEP55, which acted as a regulator in the establishment of caprine endometrial receptivity (Liu et al., 2018). Although our previous results have identified the functions of circ-8073 and miR-449a in CEECs, the effects and interactions of miR-34a/c and circ-8073 in CEECs and their role in RE establishment remain largely unknown.
In this study, we investigated the functions of miR-34a/c in CEECs and their effects on RE marker genes, and described the interactions between miR-34a/c and circ-8073/miR-449a in CEECs in vitro.
2 MATERIALS AND METHODS
2.1 Ethics statement
All experimental dairy goats in this study were maintained according to Proclamation No. 5 of the Ministry of Agriculture, P. R. China, and all animal protocols were approved by the Review Committee for the Use of Animal Subjects of Northwest A&F University.
2.2 Endometrial sample collection
Endometrial samples from dairy goats at gestational days 5 (D5, PE, n = 3) and 15 (D15, RE, n = 6) were collected from the inner wall of the uterine cavity (Y. Song et al., 2015; L. Zhang et al., 2015). A portion of the endometrial samples was immediately frozen in liquid nitrogen for total RNA extraction, whereas the remaining samples were immediately placed in phosphate buffered saline with penicillin (100 IU/ml) and streptomycin (50 mg/ml) for the isolation of CEECs.
2.3 Cell culture and treatment
Primary CEECs were isolated from the endometrial samples of dairy goats, purified, and cultured, as previously described (Lei et al., 2017; L. Zhang, Liu, Liu, Zhou, et al., 2017). The overexpression plasmids of CEP55 and circ-8073 have previously been constructed (Liu et al., 2018), and the primers are shown in Table 1. Mature miR-34a/34c/449a mimics, negative control (NC), miR-34a/34c/449a inhibitors, NC-inhibitor (NCH), CEP55-siRNA, and circ-8073-siRNA were purchased from GenePharma (Shanghai, China). To detect the treatment efficiency of the ERK1/2 inhibitor (FR-180204; Beyotime, Shanghai, China) on CEP55, the ERK1/2 inhibitor was diluted in F12 medium to achieve concentrations of 0, 5, 10, and 20 μM. CEECs were treated with either the ERK1/2 inhibitor or the protein kinase B (AKT)-inhibitor Perifosine (Beyotime, Shanghai, China) for 48 hr, and CEECs were harvested daily for western blot (WB) analysis. CEECs were plated at a density of 7.5 × 105 in six-well plates and were transfected at 50% confluency using Lipofectamine 2000 (Invitrogen, Carlsbad).
| Gene | GenBank accession no. | Primer sequences (5′→3′) |
|---|---|---|
| β-Actin | XM_018039831.1 | F: GATCTGGCACCACACCTTCT |
| R: GGGTCATCTTCTCACGGTTG | ||
| CEP55 (qPCR) | XM_005698222.3 | F: CGTGAGTTTGCCTTCACAGA |
| R: TTCATTTAACGCAGCAGTGG | ||
| CEP55 (Check2) | / | F: GCTCGAGAGGAGGAGCAAACAAGGG |
| R: ATGCGGCCGCCTCGGAAATGCAAAGCAG | ||
| CEP55 (pcDNA3.1) | / | F: ACGGGTACCATGACTTCCAGAAATCCCAAAGATTTAATTAAAAGTAAATGGGG |
| R: CGCTCGAGCTACTTGGAACAGTATTCAACGTGG | ||
| circ-8073 (qPCR) | / | F: CCGTGGGTTTGCTGACCATT |
| R: GACTCCACGAAATCGGCCTC | ||
| circ-8073 (Check2) | / | F: CGCTCGAGATGCGAGTGTCTCTCTGGCTT |
| R: GCGCGGCCGCCTTCGCATACGGGGCAGC | ||
| circ-8073 (pcD2.1-ciR) | / | F: GGGGTACCTGAAATATGCTATCTTACAGATGCGAGTGTCTCTCTGGCTT |
| R: CGGGATCCTCAAGAAAAAATATATTCACCTTCGCATACGGGGCAGC | ||
| U6 | / | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | ||
| miR-34a-5p-Loop | / | gtcgtatccagtgcagggtccgaggtattcgcactggatacgacACAACCAG |
| miR-34a-5p | / | gcgcgcTGGCAGTGTCTTAG |
| miR-34c-5p-Loop | / | gtcgtatccagtgcagggtccgaggtattcgcactggatacgacGCAATCAG |
| miR-34c-5p | / | gcgcgcAGGCAGTGTAGTTAG |
| miR-449a-Loop | / | gtcgtatccagtgcagggtccgaggtattcgcactggatacgacGCCAGCTA |
| miR-449a | / | gcgcgcTGGCAGTGTATTGT |
| Reverse Primer | / | GTGCAGGGTCCGAGGT |
- Note: The characters with underscore were restriction enzyme cutting site of xhoІ and not І for constructing psiCHECK2. The bold italicized characters with underscore were restriction enzyme cutting site of KpnI and xhoІ for constructing pcDNA3.1. The blod italicized characters with underscore were restriction enzyme cutting site of KpnI and BamHI for constructing pCD2.1-ciR. circ-8073 (qPCR) was outward-facing primer and divergently primed.
2.4 Luciferase assay
To identify the targets of miR-34a/34c, bioinformatic analysis of miRNA-binding sequences of CEP55 and circ-8073 was performed using MiRanda and Targetscan7.0. The wild-type or mutated psiCHECKTM-2 plasmids of CEP55 3′-UTR and full circ-8073 used for luciferase assays have previously been constructed (Liu et al., 2018), and the primers are shown in Table 1. psiCHECK2-WT and psiCHECK2-MUT were cotransfected with the miR-34a/34c mimics into 293T cells. The dual-luciferase assay was performed as previously described (Lei et al., 2017; L. Zhang, Liu, Liu, Zhou, et al., 2017), and each experiment was performed in triplicate.
2.5 RNA extraction and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using the TRIzol reagent (Takara, Dalian, China), according to the manufacturer's instructions. To quantify the amount of circRNA, aliquots of total RNA were incubated at 37°C with or without (mock) 3 U/μg of RNase R (Epicenter Biotechnologies, Chicago, IL) for 15 min. The purity and concentration of RNA were determined by measuring the optical density at wavelengths of 260 and 280 nm using an Epoch microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT). Total RNA (900 ng) was used to convert mRNAs into cDNAs using the PrimeScript RT Reagent Kit (Takara). RT-qPCR combined with N = was used to detect the expression levels of mRNAs and miRNAs, and β-actin and U6 were used as references. The primers used are shown in Table 1.
2.6 Protein extraction and WB analysis
Protein extraction was performed using Solarbio (Beijing, China), in accordance with the manufacturer's instructions. CEECs were transfected with miR-34a/34c/449a mimics, NC, miR-34a/34c/449a inhibitors, NCH, CEP55-siRNA, circ-8073-siRNA, pc2.1-circ-8073, or pc3.1-CEP55 for 48 hr. The cells were then harvested and lysed in RIPA buffer (Solarbio) at 4°C for 30 min and centrifuged (11,000g) for 10 min. Soluble proteins in the supernatants were collected, and total protein concentrations were determined using the Bio-Rad DC Protein Assay (Hercules, CA) after diluting with 100 μl 4 × protein loading buffer (Solarbio) and boiling for 5–8 min. As previously described (Lei et al., 2017; L. Zhang, Liu, Liu, Zhou, et al., 2017), WB analysis was performed using Quantity One software (Bio-Rad, Hercules, CA), in accordance with the manufacturer's instructions, using the antibodies listed in Table 2.
| Name | Manufacturer | Product number |
|---|---|---|
| β-Actin | Beyotime, Shanghai, China | AA128 |
| CEP55 | BBI, Shanghai, China | D198043 |
| p-CEP55 (Ser425) | Absci, United States | #AB12585 |
| FOXM1 | SAB, United States | #32671 |
| VEGF | Boster, Wnhan, China | BA0407 |
| OPN | Boster, Wnhan, China | BM0032 |
| PTGS2 | Boster, Wnhan, China | BA0738 |
| PRL | Boster, Wnhan, China | BA14521 |
| BAX | Beyotime, Shanghai, China | AB026 |
| BCL2 | Beyotime, Shanghai, China | AB112 |
| Caspase-3 | Cell Signaling, America | #9662 |
| p-PI3K p110 β (Ser1070) | Bioss, Beijing, China | bs-6417R |
| PI3K p110 β | Bioss, Beijing, China | bs-6423R |
| AKT | Cell Signaling, America | #9272 |
| p-AKT (Ser473) | Cell Signaling, America | #9271 |
| p-mTOR (S2448) | Boster, Wnhan, China | BM4840 |
| mTOR | Boster, Wnhan, China | BM4182 |
| RAS | Gene Tex, America | GTX132480 |
| p-RAF1 (Ser642) | BBI, Shanghai, China | D151385 |
| RAF1 | BBI, Shanghai, China | D220484 |
| p-MEK1 (Thr291) | BBI, Shanghai, China | D155140 |
| MEK1 | BBI, Shanghai, China | D261061 |
| p-ERK1/2 (Thr202/Tyr204) | Cell Signaling, America | #9101 |
| ERK1/2 | Cell Signaling, America | #9102 |
| HRP-labeled goat anti-rabbit IgG (H+L) | Beyotime, Shanghai, China | A0208 |
| HRP-labeled goat anti-mouse IgG (H+L) | Beyotime, Shanghai, China | A0216 |
2.7 Cell proliferation assay
The Edu (Ribobio, Guangzhou, China) and Cell Counting Kit-8 (CCK-8) assays (ZETA) were used to evaluate the proliferation of CEECs, as previously described (H. Li et al., 2017). CEECs were seeded in 96-well plates, at a density of 2 × 103 cells/well, with 100 μl of culture medium per well, and transfected with miR-34a/c mimics, miR-34a/c inhibitors, NC, or NCH. Each treatment group had six independent replicates.
2.8 Cell apoptosis and cycle assays
CEEC apoptosis was evaluated using the flow cytometry method (FCM) and the Annexin V-FITC PI staining apoptosis assay kit (Liankebio, Hangzhou, China), according to the manufacturer's instructions. Annexin V-positive and PI-negative cells were defined as early apoptotic, and Annexin V-negative and PI-positive cells were defined as apoptotic, following the manufacturer's protocol. The Cell Cycle Staining Kit with a flow cytometer (Liankebio, Hangzhou, China) was used to assess the CEEC cell cycle, as previously described (L. Zhang, Liu, Liu, Zhou, et al., 2017). Analyses were performed using a flow cytometer (BD Biosciences, San Diego, CA).
2.9 Statistical analysis
Results were expressed as mean ± standard error of the mean (SEM), and analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL). One-way analysis of variance and the least significant difference (LSD) test were used to determine differences between the test groups. Results were considered significant at *p < .05 and highly significant at **p < .01.
3 RESULTS
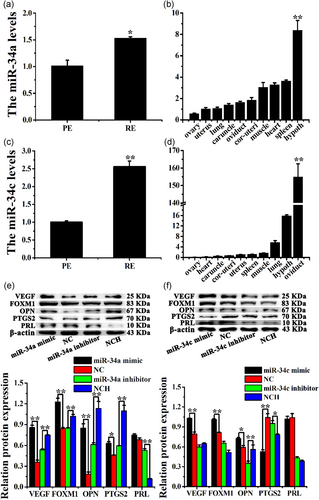
3.1 miR-34a/c levels in RE were higher than those in PE
The expression levels of miR-34a were lower in PE than in RE (Figure 1a), which was consistent with our previous sequencing results (Y. Song et al., 2015). miR-34a was widely expressed in the ovary, uterus, lung, caruncle, oviduct, cornua uterus, muscle, heart, spleen, and hypothalamus of dairy goats (Figure 1b). Moreover, the expression levels of miR-34c were markedly increased in RE compared with those in PE (Figure 1c). Furthermore, miR-34c was extensively expressed in the hypothalamus, lung, muscle, spleen, uterus, cornua uterus, caruncle, heart, and ovary (Figure 1d).
The transfection efficiencies of miR-34a/c were detected, and the results indicated that the expressions of miR-34a and miR-34c were significantly increased by the miR-34a and miR-34c mimics, respectively, in CEEC in vitro (Figure S1A,B, p < .01).
3.2 miR-34a/c regulate the expression of some marker genes of endometrial receptivity in dairy goats
Changes in vascular endothelial growth factor (VEGF), forkhead box M1 (FOXM1), PRL, prostaglandin-endoperoxide synthase 2 (PTGS2), and OPN protein levels were detected in CEECs treated with miR-34a/c mimics or inhibitors in vitro. The levels of VEGF, FOXM1, OPN, and PTGS2 were significantly upregulated by the miR-34a mimic, whereas they were significantly downregulated by the miR-34a inhibitor in CEEC in vitro (Figure 1e, p < .01). In addition, the miR-34a mimic had no effect on the PRL protein level; however, the miR-34a inhibitor increased these levels (Figure 1e, p < .01). miR-34c also significantly upregulated the protein levels of VEGF, FOXM1, and OPN, whereas the inhibitor only decreased the OPN protein levels in CEEC in vitro (Figure 1f, p < .01). In addition, the miR-34c mimic decreased the PTGS2 protein levels (p < .01), whereas the miR-34c inhibitor increased these levels (p < .05); however, miR-34c did not affect the protein levels of PRL in CEEC in vitro (Figure 1f). These results indicated that miR-34a/c may regulate the expression of VEGF, FOXM1, OPN, PTGS2, and PRL in CEEC in vitro.
3.3 miR-34a/c mimics promote CEEC apoptosis in vitro
The Edu and CCK-8 assays revealed that the miR-34a/c mimics markedly inhibited CEEC proliferation, whereas it was promoted by the miR-34a/c inhibitors (Figure 2a,b). Furthermore, the Annexin V-FITC/PI assay combined with FCM showed that the miR-34a/c mimics promoted CEEC apoptosis (Figures 2c and S3A), whereas the miR-34a/c inhibitors had no effect on the CEECs (Figure S2A and S3B). The cell-cycle assay indicated that the miR-34a/c mimics halted CEEC in the S phase (Figures 2d and S4A), whereas the miR-34a/c inhibitors had no effect on the CEEC cycle (Figures S2B and S4B).
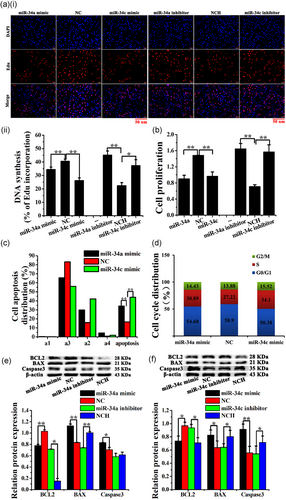
Moreover, miR-34a/c mimics significantly increased the protein levels of BAX and caspase-3 proteins, whereas they reduced the BCL2 levels. In contrast, the miR-34a/c inhibitors markedly reduced the levels of BAX and increased the levels of BCL2 in CEECs in vitro (Figure 2e,f).
3.4 miR-34a/c were sponged by circ-8073
In the present study, miR-34a and miR-34c were predicted to functionally target circ-8073 through predicted binding sites. To validate these predictions, psiCHECK2 vectors were used (Figure 3a), which demonstrated that the luciferase activity of the WT-circ-8073 + miR-34a/c mimic groups decreased significantly (p < .05). Furthermore, no change was detected in the MUT-circ-8073 +miR-34a/c mimic groups (Figure 3b,c).
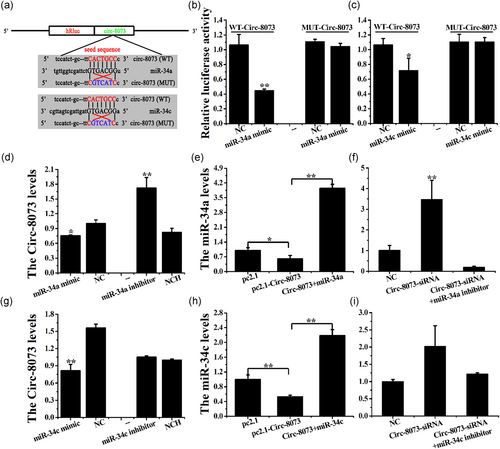
Additionally, the miR-34a mimic decreased the circ-8073 levels (p < .05), whereas the miR-34a inhibitor increased these levels in CEECs in vitro (Figure 3d, p < .01). Overexpression of circ-8073 and pc2.1-circ-8073 decreased the miR-34a levels (Figure 3e, p < .05), whereas the knockdown of circ-8073 markedly increased the miR-34a levels in CEECs in vitro (Figure 3f, p < .01). In addition, the miR-34c mimic decreased the circ-8073 levels (p < .01), whereas the miR-34c inhibitor had no effect on these levels in CEECs in vitro (Figure 3g). circ-8073 decreased the miR-34c levels (Figure 3h, p < .01), and the knockdown of circ-8073 increased these levels in CEECs in vitro (Figure 3i, p < .05). These results indicated a possible negative regulation between circ-8073 and miR-34a/c via the binding sites in CEECs in vitro.
3.5 CEP55 is a common target gene of miR-34a/c
CEP55 was predicted to be a target gene of miR-34a/c, for the predicted binding site in the 3′-UTR, using the publicly available programs TargetScan 7.0 and MiRanda. To validate this prediction, the psiCHECK-2 reporter (WT) and mutated (MUTm) plasmids were constructed (Figure 4a). The results showed that miR-34a/c significantly mimicked the luciferase activity of WT-CEP55-3′-UTR (p < .01), but did not alter the luciferase activity of MUT-CEP55-3′-UTR (Figure 4b,c).
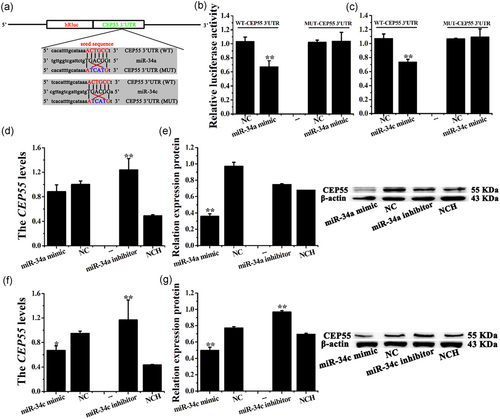
It is worth noting that the miR-34a mimic downregulated the CEP55 mRNA levels to some extent, although the difference was not significant. However, the miR-34a inhibitor dramatically increased the CEP55 mRNA levels in CEECs in vitro (Figure 4d, p < .01). The miR-34a mimic decreased the CEP55 protein levels (Figure 4e, p < .01), whereas the miR-34a inhibitor had no significant effect on CEECs in vitro. Moreover, the miR-34c mimic decreased the CEP55 mRNA (p < .05) and protein levels (p < .01), whereas the inhibitor upregulated these levels (Figure 4f,g, p < .01).
3.6 Relationship between miR-34a/c and miR-449a in CEECs
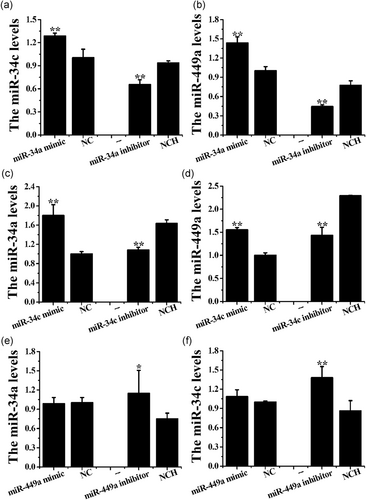
The stem-loop RT-qPCR showed that the miR-34a mimic upregulated the expression levels of miR-34c and miR-449a (p < .01), whereas the miR-34a inhibitor decreased these levels (Figure 5a,b, p < .01). In addition, the miR-34c mimic increased the expression levels of miR-34a and miR-449a, whereas the miR-34c inhibitor decreased these levels in CEEC in vitro (Figure 5c,d, p < .01). The miR-449a mimic had no significant effect on the miR-34a/c levels, whereas the miR-449a inhibitor significantly upregulated these levels in CEECs in vitro (Figure 5e,f).
3.7 miR-34a/c regulates the phosphoitide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway in CEECs
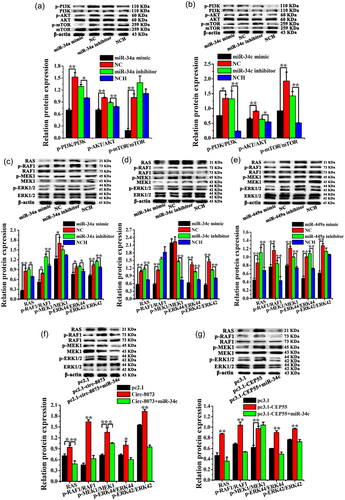
Our previous study confirmed that miR-449a inhibited CEEC proliferation by downregulating circ-8073 and CEP55 via the PI3K/AKT/mTOR pathway (Liu et al., 2018), whereas the effects of miR-34a/c on CEEC proliferation and apoptosis via the PI3K/AKT/mTOR pathway remained unknown. In the present study, the miR-34a mimic decreased the protein levels of p-PI3K/PI3K, p-AKT/AKT, and p-mTOR/mTOR (Figure 6a, p < .01). The miR-34a inhibitor markedly increased the levels of p-PI3K/PI3K and p-AKT/AKT, whereas the levels of p-mTOR/mTOR were not significantly altered in CEECs in vitro. Furthermore, the miR-34c mimic significantly reduced the levels of p-PI3K/PI3K, p-AKT/AKT, and p-mTOR/mTOR, whereas these levels were significantly increased by the miR-34c inhibitor in vitro (Figure 6b). These results indicated that miR-34a/c regulated CEEC apoptosis via the PI3K/AKT/mTOR pathway.
3.8 miR-34a/c, miR-449a, circ-8073, and CEP55 regulate the RAS/RAF/MEK/ERK pathway in CEECs
To further study the effects of miR-34a/c, circ-8073, and CEP55 on CEEC survival, the protein levels involved in the RAS/RAF/MEK/ERK pathway were measured using WB. As shown in Figure 6c, the miR-34a mimic significantly reduced the protein levels of RAS, p-RAF1/RAF1, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42. The miR-34a inhibitor markedly increased the protein levels of RAS, p-RAF1/RAF1, p-ERK44/ERK44, and p-ERK42/ERK42 in CEECs in vitro. In addition, the miR-34c mimic decreased the protein levels of RAS, p-RAF1/RAF1, p-ERK44/ERK44, and p-ERK42/ERK42 (Figure 6d, p < .01), whereas the miR-34c inhibitor increased the protein levels of RAS, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42 in CEECs in vitro (Figure 6d, p < .01). Moreover, the miR-449a mimic reduced the protein levels of RAS, p-RAF1/RAF1, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42 (Figure 6e, p < .01), and this reduction was reversed by the miR-449a inhibitor in CEECs in vitro.
circ-8073 significantly increased the protein levels of RAS, p-RAF1/RAF1, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42, whereas the circ-8073 +miR-34c group was reduced in CEECs in vitro (Figure 6f). However, the knockdown of circ-8073 had no apparent effect on the levels of p-RAF1/RAF1, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42, but it decreased the protein levels of RAS in CEECs in vitro (Figure S5A, p < .01). As expected, CEP55 increased the protein levels of RAS, p-RAF1/RAF1, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42 (Figure 6g, p < .01). Additionally, the knockdown of CEP55 markedly inhibited the expression of RAS, p-RAF1/RAF1, p-MEK1/MEK1, p-ERK44/ERK44, and p-ERK42/ERK42 in CEECs in vitro (Figure S5B). As mentioned previously, it was concluded that miR-34a, miR-34c, miR-449a, circ-8073, and CEP55 regulated CEEC apoptosis and proliferation via the RAS/RAF/MEK/ERK pathway in dairy goats.
3.9 Effects of the ERK1/2 inhibitor on CEP55 and apoptosis-associated proteins in CEECs
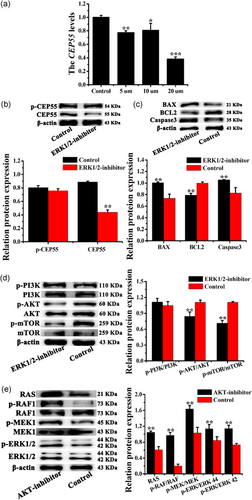
It has been reported that an ERK2 inhibitor blocks the phosphorylation of CEP55 in cancer cells (Fabbro et al., 2005). In the present study, the ERK1/2 inhibitor was diluted to different concentrations (0, 5, 10, and 20 μM) in F12 medium to investigate the effects of ERK1/2 on CEP55 expression in CEECs. The lowest CEP55 mRNA level was recorded with 20 μM ERK1/2 inhibitor concentration in CEECs; thus, this concentration was selected for further analyses. The ERK1/2 inhibitor reduced the total CEP55 level (Figure 7a, p < .01), but had no effect on the phosphorylation of CEP55 in CEECs in vitro (Figure 7b). In addition, the apoptosis-associated proteins BCL2, BAX, and caspase-3 were examined in CEECs treated with the ERK1/2 inhibitor, and the results showed that the ERK1/2 inhibitor significantly increased the levels of the antiapoptotic proteins BAX and caspase-3, whereas it reduced the BCL2 levels (Figure 7c, p < .01). Furthermore, WB was used to detect the treatment efficiency of the ERK1/2 inhibitor in CEECs. The results indicated that the phosphorylation level of ERK1/2 was significantly reduced (p < .01), and the ERK1/2 inhibitor had no effect on the total ERK1/2 protein level in CEECs in vitro (Figure S6A,B). These results indicated that the ERK1/2 inhibitor could induce CEEC apoptosis and block the RAS/RAF/MEK/ERK pathway in CEECs in vitro.
3.10 Cross-talk between the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways in CEECs
To confirm the occurrence of cross-talk between the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways in CEECs, the effects of the ERK1/2 inhibitor or AKT-inhibitor on the phosphorylation of proteins in the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways were examined. The results showed that the ERK1/2 inhibitor significantly reduced the levels of p-AKT and p-mTOR in CEECs in vitro, whereas the levels of p-PI3K remained unaffected (Figure 7d). It was interesting to note that the AKT-inhibitor increased the levels of RAS, p-RAF1, p-MEK1, and p-ERK1/2 in CEECs in vitro (Figure 7e, p < .01). These findings demonstrated positive and negative feedback relationships and cross-talk between the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways in CEECs.
4 DISCUSSION
In our previous study, the miRNA profile related to endometrial receptivity during embryo implantation in dairy goats was reported for the first time (Y. Song et al., 2015). Furthermore, miR-182 (L. Zhang, Liu, Liu, Song, et al., 2017), miR-26a (Lei et al., 2017), and miR-449a (An et al., 2017; Liu et al., 2018) were found to play important roles in the development of endometrial receptivity in dairy goats, and were more highly expressed in RE than in PE. In the present study, the RT-qPCR results showed that both miR-34a and miR-34c were significantly higher in RE than in PE, which is consistent with our previous sequencing analysis results (Y. Song et al., 2015). These findings suggest that miR-34a/c participate in RE establishment in dairy goats.
RE establishment is a complicated and dynamic process during which the endometrium undergoes morphological and physiological changes, including cell proliferation, cell apoptosis, and cell adhesion (S. Q. Cheng et al., 2009; Heng, Li, Stephens, Rainczuk, & Nie, 2010; L. Zhang, Liu, Han, et al., 2017). EEC apoptosis is an indispensable physiological event for embryo implantation (Galán et al., 2000), and studies have shown that miRNAs can induce EEC apoptosis in mice (Nie et al., 2017; Pan et al., 2018; Yuan et al., 2015). Furthermore, our previous studies have demonstrated that miR-182 and miR-449a can contribute to the development of endometrial receptivity by inducing CEEC apoptosis in dairy goats (Liu et al., 2018; L. Zhang, Liu, Liu, Zhou, et al., 2017; L. Zhang, Liu, Liu, Song, et al., 2017). Moreover, some studies have shown that miRNAs are negative regulators of the antiapoptosis gene BCL2 in cancer cells (Balatti & Croce, 2019; Pekarsky & Croce, 2019). More important, miR-34a and miR-34c can induce cell apoptosis by downregulating the antiapoptosis gene BCL2 in cancer cells (L. Li et al., 2013; R. Li, Zhang, & Zheng, 2018). In the present study, miR-34a and miR-34c were found to inhibit CEEC proliferation and induce CEEC apoptosis in dairy goats.
miR-34a, miR-34c, and miR-449a exhibit high sequence homology, and are members of the miR-34/449 family in vertebrates, encoded at three different gene loci. They share the same seed sequence, which has robust functional redundancy (Chevalier et al., 2015; Ebner & Selbach, 2014; R. Song et al., 2014). For example, centriolar protein 110 (CP110) was downregulated by miR-34/449 via the target site (R. Song et al., 2014). Lnc-OC1 represses the expression of endogenous miR-34a and miR-34c as a sponge, and vice versa (Tao, Tian, Lu, & Zhang, 2018). More important, circ-8073 functions as a sponge of miR-449a, increasing the expression of CEP55, which is a direct target of miR-449a in CEECs (Liu et al., 2018). However, it is not known if circRNA represses the expression of miR-34a/c as a sponge.
In the present study, circ-8073 exerted a sponging effect on miR-34a/c, and a negative feedback loop was observed between them in CEECs. As expected, CEP55 was a common target gene of miR-34a/c at the same seed site, and miR-34a/c also reduced the expression of CEP55 in CEECs. Interestingly, miR-34a/c actively regulated the expression of miR-449a, and positive feedback regulation was observed between miR-34a and miR-34c in CEECs in vitro. Our results indicate that circ-8073 regulates the expression levels of CEP55 by sponging miR-34a, miR-34c, and miR-449a as a ceRNA in CEECs in vitro.
The PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways are two key hyper-activated signaling pathways involved in the regulation of cell proliferation, apoptosis, growth, and survival (Butler et al., 2017; Chen et al., 2014; McCubrey et al., 2011). More important, cross-talk between the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways has been shown to regulate cell survival in human breast cancer cell lines (Moelling, Schad, Bosse, Zimmermann, & Schweneker, 2002). Furthermore, miR-34a inhibits cell proliferation by regulating the RAS/RAF/MEK/ERK signaling pathway in mesangial cells and human breast cancer cells (Chen et al., 2014; Xiao et al., 2014), and it is involved in the regulation of cell proliferation and apoptosis via the PI3K/AKT signaling pathway in gastric cancer cells (C. Cheng, Qin, Zhi, Wang, & Qin, 2017). Moreover, Chevalier et al. reported that miR-34/449 can inhibit the activity of RAS (Chevalier et al., 2015). However, the effect of miR-34a/c on cell proliferation and apoptosis via the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways has not been studied. In the present study, miR-34a/c were shown to inhibit the protein phosphorylation activity of both PI3K/AKT/mTOR and RAS/RAF/MEK/ERK signaling pathways.
The effects of circ-8073, miR-449a, and CEP55 on the RAS/RAF/MEK/ERK pathway in CEECs were also investigated, as our previous study only confirmed that miR-449a inhibited CEEC proliferation by downregulating circ-8073 and CEP55 via the PI3K/AKT/mTOR pathway (Liu et al., 2018). In the present study, circ-8073 and CEP55 were shown to elevate protein phosphorylation levels of the RAS/RAF/MEK/ERK pathway, whereas miR-449a was shown to reduce this activity in CEECs in vitro. These results confirmed that miR-34a/c/449a induced CEEC apoptosis by downregulating circ-8073 and CEP55 through the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways.
In addition, the effects of miR-34a/c on various biomarkers of endometrial receptivity, including VEGF (Halder et al., 2015), OPN (Coutifaris, Omigbodun, Ziolkiewicz, & Hoyer, 1999; Qi, Xie, Liu, & Zhou, 2014), PTGS2 (Achache & Revel, 2006), and PRL (Garzia et al., 2004; Negami & Tominaga, 1991), were investigated in CEECs in vitro. Our previous study demonstrated that FOXM1 upregulated the expression of VEGF in CEECs, and that the knockdown of CEP55 might contribute to the development of endometrial receptivity by regulating FOXM1 and VEGF in dairy goats (Liu et al., 2018). FOXM1, which is a transcription factor (Wierstra & Alves, 2007), can be upregulated by OPN in human uterine epithelial cells, and may play an important role in the formation of RE during embryo implantation (Xie, Li, & Kong, 2014). In the present study, miR-34a/c significantly increased the VEGF, FOXM1, and OPN levels, whereas they had no effect on PRL in CEECs. PRL is induced in endometrial stromal cells during decidualization, and it has been recognized as a marker of decidualization (Fluhr et al., 2006; Kimura et al., 2001; D. F. Wang et al., 2007; Q. Zhang et al., 2012). However, endometrial decidualization during embryo implantation has not been studied in dairy goats. Our results indicated that miR-34a/c could contribute to the development of RE by regulating VEGF, FOXM1, and OPN in vitro.
ERK1/2 are important members of the mitogen-activated protein kinase (MAPK) conserved cascade, which are involved in the regulation of cell proliferation, apoptosis, cell-cycle progression, and gene expression, through phosphorylated downstream proteins (Cerioni & Cantoni, 2010; White, Stevens, Haidekker, & Frangos, 2005). ERK2-inhibition blocks the phosphorylation of CEP55 in cancer cells (Fabbro et al., 2005). In the present study, the ERK1/2 inhibitor reduced the total protein levels of CEP55, but did not affect its phosphorylation in CEECs in vitro. Furthermore, our results indicated that the ERK1/2 inhibitor reduced CEEC apoptosis by regulating the expression of BCL2, BAX, and caspase-3, and blocked the RAS/RAF/MEK/ERK pathway in CEECs in vitro. Hence, we speculated that the RAS/RAF/MEK/ERK pathway might play a vital role in RE development in dairy goats.
The PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways are regulated by cross-talks and feedback loops (Guenther, Graab, & Fulda, 2013). Studies have shown that RAS can activate these pathways (Asati, Mahapatra, & Bharti, 2016; Mccubrey et al., 2011), and that the balance between cell arrest and proliferation is regulated by positive feedback from p-AKT-ERK (Yaman, 2016). Furthermore, mTOR is a serine-threonine protein kinase that regulates cell growth, proliferation, and apoptosis, and studies have shown that it can be stimulated by RAS signaling in cancer cells (Jhanwar-Uniyal et al., 2017, 2019). Moreover, AKT phosphorylation is activated by the MEK-inhibitor, whereas the dual PI3K/mTOR-inhibitor enhances ERK phosphorylation (Guenther et al., 2013). Our previous study also confirmed that the AKT-inhibitor not only induced CEEC apoptosis, but also blocked the PI3K/AKT/mTOR signaling pathway (Liu et al., 2018). Therefore, the effects of the ERK1/2 inhibitor or the AKT-inhibitor on phosphorylation during the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways were examined to confirm the presence of cross-talk between these two pathways in CEECs in vitro. In the present study, positive and negative feedback loops and cross-talk between the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK signaling pathways were detected. For example, the ERK1/2 inhibitor blocked the AKT/mTOR signaling pathway, whereas the RAS/RAF/MEK/ERK signaling pathway was activated by the AKT-inhibitor in CEECs in vitro.
Taking these findings and our previous study results (Liu et al., 2018) into consideration, we conclude that miR-34a/34c/449a not only induce apoptosis in CEECs by downregulating circ-8073 and CEP55 via the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways, but also contribute to the establishment of endometrial receptivity in dairy goats (Figure 8).

ACKNOWLEDGMENTS
This study was supported by PhD research startup foundation of Northwest A&F University (00400/Z109021811), China Postdoctoral Science Foundation (2019M653776), Natural Science Foundation of Shaanxi Province (2020JQ-265), and the National Key Research and Development Program of China (2016YFD0500508). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
X. R. L. and L. Z. conceived and designed the experiments; X. R. L., S. C. C., J. Z. C., Y. L. C., Y. X. L., J. C. H., and X. P. A. performed the experiments; X. R. L. analyzed the data; L. Z., B. Y. C., and Y. X. S. contributed reagents/materials/analysis tools; X. R. L. wrote the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon request.