Blood and tissue levels of lncRNAs in periodontitis
Arezou Sayad and Soudeh Ghafouri-Fard contributed equally.
Abstract
Periodontitis is a complex disorder that affects a large number of human beings from different ethnic groups. This condition has been associated with dysregulation of a number of genes, among them are long noncoding RNAs (lncRNAs). In the current study, we assessed the expression of four lncRNAs (BDNF-AS, MIAT, MIR137HG, and PNKY) as well as BDNF in the peripheral blood and gingival tissues obtained from patients with periodontitis and healthy subjects. The expression of BDNF was significantly lower in blood samples of male patients with periodontitis compared with male controls (posterior β of RE = −4.754, p = .048). However, there was no significant difference in the expression of BDNF in tissue samples from the cases and controls. The expression of BDNF-AS was significantly lower in the tissue samples of patients compared with control tissue samples (posterior β of RE = −2.151, p = .019). Such an expression difference was detected between male subgroups as well (posterior β of RE = −3.679, p = .009). However, expression of this lncRNA was not different in blood samples obtained from patients compared with healthy subjects. The expression of PNKY was significantly higher in tissue samples obtained from female patients compared with sex-matched controls (posterior β of RE = 6.23, p = .037). Blood levels of this lncRNA were not different between cases and controls. There was no significant difference either in the tissue expression or in blood expression of MIR137HG or MIAT between cases and controls. The current study indicates the putative role of BDNF, BDNF-AS, and PNKY in the pathophysiology of periodontitis and potentiates these genes as candidates for functional studies.
1 INTRODUCTION
Periodontitis is a condition caused by an intricate interaction between immune factors, microbial content of the oral cavity, and lifestyle practices (Berezow & Darveau, 2011). Genetic factors have prominent roles in the regulation of this interplay (Schaefer, 2018). A group of transcripts with sizes more than 200 nucleotides and an inability to be translated into proteins have been recently recognized to regulate immune responses against bacteria (Hadjicharalambous & Lindsay, 2019). Thus, these so-called long noncoding RNAs (lncRNAs) might be involved in the pathophysiology of periodontitis (Li et al., 2018). We have recently reported dysregulation of UCA1, NEAT1, FAS-AS1, and NKILA lncRNAs in peripheral blood or gingival tissues of patients with periodontitis compared with healthy subjects (Sayad et al., 2020). In the current study, we have selected four other lncRNAs to assess their expression in these two sets of samples. The basis for selection of these lncRNAs was their putative role in the regulation of immune responses. Brain-derived neurotrophic factor-antisense RNA (BDNF-AS) is a natural antisense transcript that is transcribed opposite the BDNF gene and regulates expression of the sense transcript (Zhong et al., 2017). In addition to the well-appreciated role of BDNF in survival, differentiation, and neurite outgrowth, this factor has essential roles in the regulation of immune responses. BDNF has been shown to decrease quantities of natural killer (NK) cells but enhance accumulation of natural killer T cells in the spleen of xenotransplanted mice. These functions possibly suppress rejection of the peripheral nerve following xenotransplantation (Yu et al., 2016). More recently, BDNF has been shown to regulate the neuro-immune axis (Jin, Sun, Yang, Cui, & Xu, 2019). This speculation is supported by the observed decrease in the BDNF level after administration of pro-inflammatory cytokines or lipopolysaccharide (LPS; Frühauf-Perez et al., 2018). Moreover, IFN-α has been shown to reduce systemic BDNF levels (Lotrich, Albusaysi, & Ferrell, 2013). The lncRNA MIAT has been recognized as an essential regulator of cell invasion, migration, and proliferation which exerts its effects through modulation of the PI3K/AKT signaling pathway (Jin et al., 2019). Notably, this pathway participates in the regulation of interleukin-10 (IL-10) and IL-12 by Porphyromonas gingivalis LPS as one of the main etiological factors in periodontitis (Martin et al., 2003). MIR137HG is a host gene for miR-137 which has been identified as a novel biomarker for multiple sclerosis (MS) disorder (Ehya, Abdul Tehrani, Garshasbi, & Nabavi, 2017). The lncRNA PNKY is a regulator of cortical development (Andersen et al., 2019). This lncRNA controls neuronal differentiation of embryonic and postnatal neural stem cells through interaction with the splicing regulator polypyrimidine tract-binding protein 1 (PTBP1; Ramos et al., 2015). This RNA binding protein has a crucial role in B cell selection in germinal centers of secondary lymphoid tissues (Monzón-Casanova et al., 2018). Thus, it is possible that PNKY participates in the regulation of immune reactions. In the current case-control study, we examined expression of BDNF, BDNF-AS, MIAT, MIR137HG, and PNKY in blood samples and gingival tissues obtained from patients with periodontitis and healthy individuals.
2 MATERIALS AND METHODS
2.1 Study participants
Periodontitis tissue samples were obtained during periodontal flap surgeries from patients. Control tissues were excised through crown lengthening or implant surgery from individuals that did not have periodontitis. Totally, 30 tissue samples from patients and 30 control tissue samples in addition to 23 blood samples from patients and 18 healthy controls were assessed. Informed consent was obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (Ethic Code: IR.SBMU.DRC.REC.1398.090). Both clinical and radiographic criteria were evaluated for appropriate diagnosis. Inclusion criteria for the periodontitis group were the presence of at least 16 teeth, age of 18 or older, chronic periodontitis (stages II to IV) with at least two remaining periodontal pockets in each sextant after nonsurgical periodontal treatment, probing depth of 5 mm or greater, bleeding on probing (BOP), and at least 3 mm of attachment loss needing surgical periodontal treatment (Gholami et al., 2020). Exclusion criteria were cigarette smoking, systemic disorders, history of antibiotic or anti-inflammatory drug intake 3 months before surgery, pregnancy, and breastfeeding. Control samples were obtained from healthy individuals with no evidence of gingivitis or periodontitis. The existence of periodontitis was ruled out in these persons by a periodontal specialist. None of the cases or controls had a recent infection, systemic disorder, or history of maxillofacial surgery or radio/chemotherapy.
2.2 Expression assays
The hybrid-RTM blood RNA extraction kit (GeneAll, Seoul, South Korea) was used for RNA extraction from blood and tissue samples. Then, cDNA was produced using the OneStep RT-PCR Series Kit (BioFact™, Seoul, South Korea). Relative expression (RE) of the mentioned genes was quantified in all blood and tissue samples using the RealQ Plus 2x PCR Master Mix Green without ROX™ PCR Master Mix (Ampliqon, Odense, Denmark). Reactions were performed in a StepOnePlus™ RealTime PCR equipment (Applied Biosystems, Foster, CA). All reactions were prepared in duplicate. The B2M gene was used as the normalizer. Table 1 shows the sequences of the primers.
| Name | Type | Primer sequence | Primer length | PCR product length |
|---|---|---|---|---|
| B2M-F | mRNA | AGATGAGTATGCCTGCCGTG | 20 | 105 |
| B2M-R | GCGGCATCTTCAAACCTCCA | 20 | ||
| BDNF-F | mRNA | GATGCTGCAAACATGTCCATGAG | 23 | 109 |
| BDNF-R | TTTTGTCTGCCGCCGTTACC | 20 | ||
| BDNF-AS-F | lncRNA | GTGGGTCCATTCCGTGTGTG | 20 | 97 |
| BDNF-AS-R | AGCTGGTGCAGGTATCAGATTAG | 23 | ||
| MIAT-F | lncRNA | CTCACACCTCCTATTCCT | 18 | 169 |
| MIAT-R | ATCACTATCTACTTCACCAAC | 21 | ||
| PNKY-F | lncRNA | TCACTCGCTAGGACAATGGC | 20 | 120 |
| PNKY-R | TCTCCTTACAGGTGCCTCCA | 20 | ||
| MIR137HG-F | lncRNA | GGCACTCAATCATACCAC | 18 | 188 |
| MIR137HG-R | GACCATAAATCCATAACTCG | 20 |
2.3 Statistical methods
Expression levels of BDNF, BDNF-AS, MIAT, MIR137HG, and PNKY were compared between patients and controls using the Bayesian regression model. The RE level was described as Ln(Efficiency^∆Ct). The effects of independent variables were adjusted. Statistical analyses were performed in R 3.6.2 software with Rstan, ggplot 2 packages. Non-parametric quantile regression was performed using the Bootstrap method and 100 iterations. Correlations between expressions of lncRNAs were evaluated through calculation of Spearman correlation coefficients.
3 RESULTS
3.1 General information of study participants
Demographic data of cases and controls are shown in Table 2.
| Periodontitis patients | Healthy controls | |
|---|---|---|
| Total number of tissue samples | 30 | 30 |
| Female | 19 | 14 |
| Male | 11 | 16 |
| Age (mean ± SD) | 41.28 ± 2.7 | 38.8 ± 1.9 |
| Total number of blood samples | 23 | 18 |
| Female | 15 | 11 |
| Male | 8 | 7 |
| Age (mean ± SD) | 39.65 ± 3.1 | 37.91 ± 2.8 |
3.2 Expression assays
Figures 1 and 2 show RE of genes in blood and tissue samples of cases and controls, respectively.
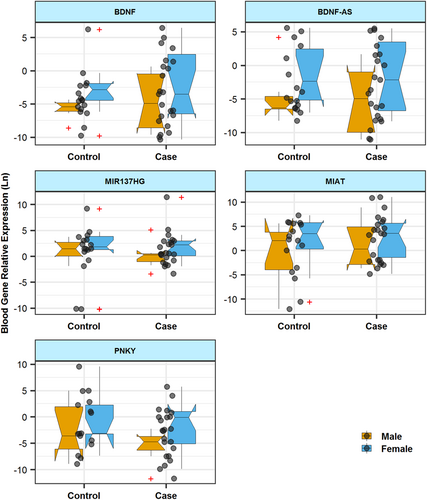
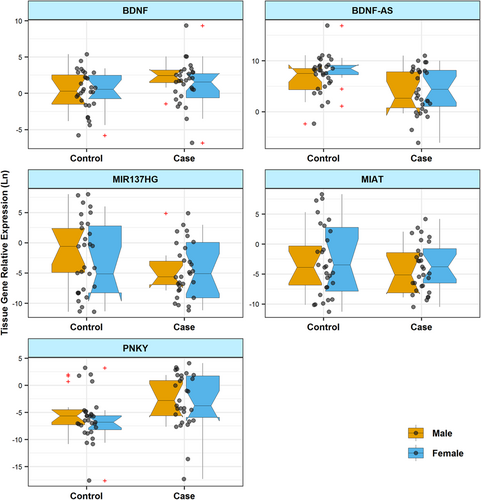
Expression of BDNF was significantly lower in blood samples of male patients with periodontitis compared with male controls (posterior β of RE = −4.754, p = .048). However, there was no significant difference in the expression of BDNF in tissue samples from cases and controls. Tissues and blood expression levels of this gene were correlated with the age of total participants. Table 3 shows the detailed data about RE of BDNF in study participants.
| Tissue | Blood | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior β of RE | SE | p Value | 95% CrI for RE | Posterior β of RE | SE | p Value | 95% CrI for RE |
| Total | Case/control | 0.628 | 0.55 | .652 | [−0.5, 1.65] | −3.101 | 1.26 | .046 | [−5.42, −0.33] |
| Gender (F/M) | −0.226 | 0.49 | .775 | [−1.18, 0.78] | 1.569 | 1.05 | .169 | [−0.57, 3.65] | |
| Age (year) | 0.051 | 0.02 | .014 | [0.005, 0.1] | 0.219 | 0.08 | .019 | [0.04, 0.38] | |
| Male | Case/control | 0.695 | 0.76 | .649 | [−0.86, 2.17] | −4.754 | 1.89 | .048 | [−8.25, −0.67] |
| Age (year) | 0.07 | 0.03 | .033 | [0.01, 0.14] | 0.343 | 0.08 | .007 | [0.16, 0.5] | |
| Female | Case/control | 0.363 | 0.82 | .55 | [−1.33, 1.92] | −1.66 | 2.39 | .974 | [−5.74, 3.9] |
| Age (year) | 0.017 | 0.04 | .324 | [−0.06, 0.11] | 0.07 | 0.18 | .958 | [−0.29, 0.4] | |
- Note. Bold values are statistically significant at p < .05.
- Abbreviations: CrI, credible interval; RE, relative expression; SE, standard error.
Expression of BDNF-AS was significantly lower in tissue samples of patients compared with control tissue samples (posterior β of RE = −2.151, p = .019). Such an expression difference was detected between male subgroups as well (posterior β of RE = −3.679, p = .009). However, expression of this lncRNA was not different in blood samples obtained from patients compared with healthy subjects (Table 4).
| Tissue | Blood | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior β of RE | SE | p Value | 95% CrI for RE | Posterior β of RE | SE | p Value | 95% CrI for RE |
| Total | Case/control | −2.151 | 1.75 | .019 | [−5.67, 0.35] | −0.853 | 1.1 | .999 | [−3.54, 0.83] |
| Gender (F/M) | 0.771 | 0.75 | .219 | [−0.74, 2.31] | 0.582 | 0.7 | .159 | [−0.54, 2.21] | |
| Age (year) | −0.003 | 0.04 | .549 | [−0.08, 0.07] | 0.02 | 0.03 | .73 | [−0.03, 0.08] | |
| Male | Case/control | −3.679 | 2.13 | .009 | [−7.14, 0.97] | −0.166 | 1.38 | >.999 | [−4.33, 2.05] |
| Age (year) | 0.024 | 0.06 | .872 | [−0.09, 0.15] | 0.015 | 0.03 | .643 | [−0.04, 0.1] | |
| Female | Case/control | −1.691 | 1.65 | .074 | [−5.85, 0.68] | −1.588 | 1.21 | .98 | [−3.65, 0.91] |
| Age (year) | −0.021 | 0.05 | .938 | [−0.12, 0.08] | 0.026 | 0.05 | .515 | [−0.08, 0.14] | |
- Note. Bold values are statistically significant at p < .05.
- Abbreviations: CrI, credible interval; RE, relative expression; SE, standard error.
There was no significant difference either in tissue expression or in blood expression of MIR137HG or MIAT between cases and controls. Expressions of these lncRNAs were not correlated with the age of either subgroup (Tables 5 and 6).
| Tissue | Blood | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior β of RE | SE | p Value | 95% CrI for RE | Posterior β of RE | SE | p Value | 95% CrI for RE |
| Total | Case/control | −1.465 | 1.05 | .444 | [−3.33, 0.76] | −1.72 | 0.97 | .443 | [−3.75, 0.1] |
| Gender (F/M) | −1.762 | 1.15 | .506 | [−3.82, 0.52] | 1.825 | 0.9 | .055 | [−0.05, 3.53] | |
| Age (year) | −0.001 | 0.06 | .095 | [−0.12, 0.11] | 0.118 | 0.06 | .05 | [0.001, 0.23] | |
| Male | Case/control | −1.327 | 1.61 | .36 | [−4.39, 2.11] | −0.924 | 0.48 | .157 | [−1.89, 0.03] |
| Age (year) | −0.056 | 0.09 | .157 | [−0.24, 0.1] | 0.004 | 0.01 | .589 | [−0.02, 0.04] | |
| Female | Case/control | −1.273 | 1.6 | .862 | [−4.23, 2.13] | −0.219 | 0.75 | .826 | [−1.76, 1.17] |
| Age (year) | 0.015 | 0.09 | .674 | [−0.18, 0.19] | 0.041 | 0.04 | .248 | [−0.02, 0.13] | |
- Abbreviations: CrI, credible interval; RE, relative expression; SE, standard error.
| Tissue | Blood | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior β of RE | SE | p Value | 95% CrI for RE | Posterior β of RE | SE | p Value | 95% CrI for RE |
| Total | Case/control | −1.033 | 1.1 | .0853 | [−3, 1.28] | 0.432 | 0.64 | .934 | [−0.69, 1.83] |
| Gender (F/M) | 0.704 | 1.06 | .706 | [−1.36, 2.73] | 0.282 | 0.49 | .56 | [−0.59, 1.36] | |
| Age (year) | −0.002 | 0.05 | .868 | [−0.1, 0.09] | −0.004 | 0.02 | .699 | [−0.05, 0.04] | |
| Male | Case/control | −1.215 | 1.94 | .867 | [−4.68, 2.58] | 0.672 | 1.22 | .876 | [−1.53, 3.43] |
| Age (year) | −0.014 | 0.07 | .835 | [−0.16, 0.12] | −0.01 | 0.04 | .959 | [−0.09, 0.06] | |
| Female | Case/control | −0.362 | 1.57 | .623 | [−3.17, 2.91] | 0.321 | 0.98 | .689 | [−1.71, 2.25] |
| Age (year) | 0.019 | 0.09 | .594 | [−0.18, 0.2] | 0.007 | 0.05 | .232 | [−0.08, 0.12] | |
- Abbreviations: CrI, credible interval; RE, relative expression; SE, standard error.
Expression of PNKY was significantly higher in tissue samples obtained from female patients compared with sex-matched controls (posterior β of RE = 6.23, p = .037). Blood levels of this lncRNA were not different between cases and controls (Table 7).
| Tissue | Blood | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters and groups | Variable | Posterior β of RE | SE | p Value | 95% CrI for RE | Posterior β of RE | SE | p Value | 95% CrI for RE |
| Total | Case/control | 3.147 | 1.77 | .034 | [0.39, 6.86] | −1.864 | 2.27 | .541 | [−6.33, 2.4] |
| Gender (F/M) | −0.378 | 0.75 | .506 | [−1.81, 1.12] | 3.887 | 1.66 | .046 | [0.39, 7.09] | |
| Age (year) | 0.04 | 0.04 | .328 | [−0.04, 0.13] | 0.062 | 0.11 | .998 | [−0.17, 0.27] | |
| Male | Case/control | 1.33 | 1.98 | .097 | [−2.09, 5.11] | −1.183 | 0.9 | .804 | [−3.07, 0.45] |
| Age (year) | 0.085 | 0.06 | .361 | [−0.02, 0.2] | 0.018 | 0.03 | .999 | [−0.03, 0.08] | |
| Female | Case/control | 6.231 | 2.02 | .037 | [1.1, 8.91] | 0.891 | 1.07 | .336 | [−0.98, 3.09] |
| Age (year) | −0.025 | 0.06 | .158 | [−0.14, 0.1] | −0.018 | 0.04 | .993 | [−0.11, 0.06] | |
- Note. Bold values are statistically significant at p < .05.
- Abbreviations: CrI, credible interval; RE, relative expression; SE, standard error.
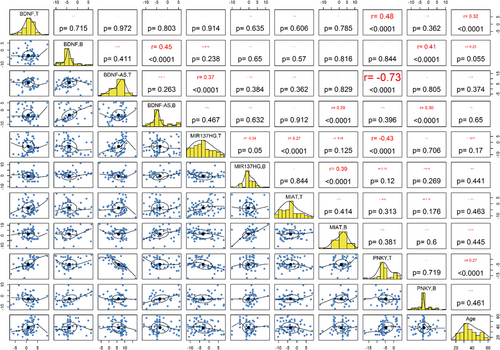
Tissue expression levels of BDNF and PNKY were correlated with age (p < .0001). Significant inverse correlations were detected between tissue expression levels of PNKY and both BDNF-AS and MIR137HG (r = −.73, p < .0001 and r = −.43, p < .0001, respectively). Significant pairwise correlations were found between blood levels of BDNF and both BDNF-AS and PNKY, blood levels of MIR137HG and MIAT as well as tissue levels of BDNF-AS and MIR137HG (Figure 3).
4 DISCUSSION
lncRNAs as important regulators of gene expression have prominent roles in regulation of immune reactions to pathogens (Hadjicharalambous & Lindsay, 2019). Based on the significance of interactions between immune factors and microbiota in the pathogenesis of periodontitis (Berezow & Darveau, 2011), lncRNAs might influence pathogenic events in this condition. A previous microarray-based analysis of lncRNA signature in periodontitis revealed differential expression of hundreds of lncRNAs between affected and adjacent normal tissues (Zou et al., 2015). In the current study, we assessed the expression of four lncRNAs and one mRNA in the peripheral blood and gingival tissues obtained from patients with periodontitis and healthy subjects. We reported a lower expression of BDNF in blood samples of male patients with periodontitis compared with male controls. However, there was no significant difference in the expression of BDNF in tissue samples from cases and controls. Corrêa et al. have previously evaluated the expression of BDNF protein in tissue samples from patients with chronic periodontitis and healthy subjects. They reported higher protein levels of this factor in affected tissues compared with normal tissues. Yet, they did not detect any association between BDNF levels and clinical factors (Corrêa et al., 2014). The inconsistency between our results and the Corrêa et al. study is best explained by the fact that we assessed mRNA levels, yet evaluated the protein levels of BDNF. This is also a clear limitation of our study, as mRNA levels are an indirect indication of protein regulation and function. BDNF has been shown to increase the expression of the anti-inflammatory cytokine IL-10, while decreasing the expression of tumor necrosis factor-α (TNF-α; Jiang et al., 2010). In a previous assay, Górska et al have demonstrated a higher frequency of IL-10 expression in healthy gingival tissues compared with tissues obtained from periodontitis patients. Moreover, they reported higher levels of TNF-α in tissue biopsies from periodontitis patients in correlation with the clinical severity of periodontitis (Gorska et al., 2003). Although we could not detect any difference in tissue expression of BDNF between cases and controls, lower blood levels of this factor in patients is in accordance with the proposed role of BDNF in the regulation of IL-10 and TNF-α and the tissue pattern of these cytokines in periodontitis. In other words, a lower level of BDNF in blood samples of patients with periodontitis is expected to result in lower concentrations of IL-10, while a higher concentration of TNF-α. Based on the anti-inflammatory role of the former and the pro-inflammatory role of the latter, decreased BDNF levels might aggravate the periodontitis course. However, this speculation should be verified through assessment of protein levels of IL-10 and TNF-α. Notably, we detected a correlation between expression levels of this gene and the age of total participants, which is in accordance with the role of BDNF in age-related pathologies (Erickson et al., 2010).
Besides this, we demonstrated lower levels of BDNF-AS in tissue samples of patients compared with control tissue samples. Such an expression difference was detected between male subgroups as well. However, expression of this lncRNA was not different in blood samples obtained from patients compared with healthy subjects. Zhong et al. (2017) have shown that BDNF-AS reduces a certain type of nerve cell apoptosis via the BDNF-TrkB-PI3K/Akt signaling pathway. Based on the acknowledged role of AKT signaling in periodontitis (Martin et al., 2003), BDNF-AS might participate in this process via modulation of the AKT pathway. It is worth mentioning that BDNF-AS has been regarded as a negative regulator of BDNF exon IX expression in neurons (Modarresi et al., 2012). The observed differences in BDNF-AS levels in tissue samples of cases and controls in spite of similar levels of BDNF between these two sets of samples might indicate a tissue-specific role of BDNF-AS. However, this speculation should be verified through functional analysis.
Moreover, we detected higher levels of PNKY in tissue samples obtained from female patients compared with sex-matched controls. The possible explanation for involvement of this lncRNA in the pathobiology of periodontitis is its interaction with the splicing regulator PTBP1 (Ramos et al., 2015) and the role of this factor in B cell selection in germinal centers of secondary lymphoid tissues (Monzón-Casanova et al., 2018). Thus, upregulation of PNKY might affect the process of B cell selection in lymphoid tissues, leading to dysregulated humoral responses in the context of periodontitis. Abnormal humoral responses have been demonstrated in patients with periodontitis for a long time since Doty, Lopatin, Syed, and Smith (1982) reported low antibacterial serum antibody titers of the immunoglobulin G (IgG) and IgA classes. Later, Kinane, Lappin, Koulouri, and Buckley (1999) demonstrated local production of IgM, and IgG and IgA subclass proteins and J-chain in the periodontitis tissues. Thus, humoral responses implicated in the pathogenesis of periodontitis, and PNKY might be involved in this process. Future studies are needed to elaborate on the underlying mechanism of this observation.
There was no significant difference either in tissue expression or in blood expression of MIR137HG or MIAT between cases and controls. These negative findings show a lack of functional relevance between these lncRNAs and periodontitis. However, we reported significant inverse correlations between tissue expression levels of PNKY and both BDNF-AS and MIR137HG. In addition, significant pairwise correlations were found between blood levels of BDNF and both BDNF-AS and PNKY, blood levels of MIR137HG and MIAT as well as tissue levels of BDNF-AS and MIR137HG. These findings indicate the presence of delicate regulatory mechanisms for lncRNA tuning which depends on the tissue context. Identification of this regulatory mechanism is a prerequisite for design of a targeted treatment modality against periodontitis. Taken together, the current study indicates the putative role of BDNF, BDNF-AS, and PNKY in the pathophysiology of periodontitis and potentiates these genes as candidates for functional studies.
ACKNOWLEDGMENT
The current study was supported by a grant from the Dental Research Center of Shahid Beheshti University of Medical Sciences.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
M. T. and S. G. F. wrote the draft and revised it. S. A. J. analyzed the data. A. S. and L. G. performed the experiment. B. S. collected the samples and corresponding data.
Open Research
DATA AVAILABILITY STATEMENT
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.