LINC00707 promotes hepatocellular carcinoma progression through activating ERK/JNK/AKT pathway signaling pathway
Abstract
Increasing evidence has demonstrated that abnormal expression of lncRNA is correlated with various malignant tumors, including hepatocellular carcinoma (HCC). Our current study was aimed to investigate the role of LINC00707 in HCC development. We observed that LINC00707 was upregulated in HCC cell lines compared with normal liver cell lines. Then, Hep3B cells and SNU449 cells were infected with LV-shLINC00707 and LV-LINC00707. LINC00707 silencing could greatly repress the proliferation and colony formation of HCC cells in vitro. On the contrary, overexpression of LINC00707 induced HCC cell proliferation and colony formation. In addition, HCC cell apoptosis was significantly enhanced and HCC cell cycle was blocked in G1 phase by LV-shLINC00707. Hep3B cells and SNU449 cell invasion capacity was restrained by the knockdown of LINC00707, whereas upregulation of LINC00707 exhibited an opposite phenomenon. Accumulating evidence has reported that ERK/JNK/AKT signaling is involved in multiple cancers, including HCC. Here, in our study, we identified that ERK/JNK/AKT signaling was dramatically restrained by silencing of LINC00707 while activated by LV-LINC00707 in HCC cells. Subsequently, an in vivo experiment was conducted, and it demonstrated that LINC00707 could modulate HCC development through activating ERK/JNK/AKT signaling. Taking the above results together, it was implied in our study that LINC00707 contributed to HCC progression through modulating the ERK/JNK/AKT pathway.
1 INTRODUCTION
Hepatocellular carcinoma (H(C) remains the fifth most frequent cancer and the third leading cause of cancer-related deaths worldwide (Bruix, Gores, & Mazzaferro, 2014; Hong et al., 2016). The 5-year survival rate of HCC patients is approximately 30% (Maluccio & Covey, 2012; Rahimi & Trotter, 2015). Although a number of advances have been made in therapeutic strategies, the prognosis for HCC remains poor because of the high rate of metastasis and recurrence (Forner, Hessheimer, Isabel Real, & Bruix, 2006; Guglielmi et al., 2014; Ishizawa et al., 2008). The underlying mechanisms of HCC are still not well understood.
The human transcriptome includes many types of RNAs, including non-coding RNAs (ncRNAs; Ponting, Oliver, & Reik, 2009). MicroRNAs are small ncRNAs that can inhibit mRNA translation through targeting the 3′-untranslated region of mRNAs (Bushati & Cohen, 2007). In addition, ncRNAs with more than 200 nucleotides are regarded as long non-conding RNA (lncRNAs; M. Fu et al., 2016; F. Zhang, Zhang, & Zhang, 2016). Previous studies have shown that lncRNAs can perform multiple physiological biological functions (Beermann, Piccoli, Viereck, & Thum, 2016). Dysregulated lncRNA expression is observed in various cancers, including lung cancer, gastric cancer, and HCC (Cao et al., 2015; Terashima, Tange, Ishimura, & Suzuki, 2017; Y. Zhao et al., 2018). LncRNAs can function through epigenetic silencing, interacting with miRNA and protein (Huang, Zheng, Hu, & Wang, 2014). Although several lncRNAs have been reported, the association of LINC00707 with HCC development and progression remains uninvestigated.
ERK/JNK/AKT signaling can play crucial roles in many cellular processes, such as cell proliferation, migration and invasion (Chang & Karin, 2001; X. Wang et al., 2012). The activation of ERK is able to promote cell proliferation in cancers (Kinkade et al., 2008; Moro, Arbini, Marra, & Greco, 2007). For instance, lncRNA Igf2as can control HCC progression through ERK signaling (Bao et al., 2017). LncRNA HCG11 can suppress the apoptosis of HCC cells via ERK/JNK/AKT signaling transduction (Y. Xu, Zheng, Liu, & Li, 2017).
In our current study, the biological roles of LINC00707 in HCC progression was concentrated on. LINC00707 was remarkably increased in HCC cells. In addition, knockdown of LINC00707 was able to inhibit HCC development in vitro while overexpression promoted HCC progression greatly. Meanwhile, the ERK/MAPK signaling pathway participated in HCC development. Hence, it was hypothesized that knockdown of LINC00707 could repress HCC development through targeting the ERK/MAPK pathway.
2 MATERIALS AND METHODS
2.1 Cell culture
Hep3B, HepG2, MHCC97L, SNU449, Huh-7 and LO2 cells were obtained from American Type Culture Collection (Manassas, VA). The cells were incubated in RPMI 1640 medium added with 10% heat-inactivated FBS (GIBCO, Carlsbad, CA), 100U/ml penicillin, and 100 mg/ml streptomycin added. The cells were incubated in an incubator with 5% CO2 at 37°C.
2.2 Lentiviral vector infection
Full-length cDNA of human LINC00707 were amplified from the mRNA of HCC cells. The sequences were designed with shLuc as the negative control, and objective products were subcloned into pcDNA3.1 (Invitrogen, Carlsbad, CA). The cells with stable expression of LINC00707 were maintained in a medium containing 800 μg/ml G418 (Sigma, St. Louis, MO).
2.3 Cell counting kit 8 assay
The cells in logarithmic growth phase were digested using trypsin. The cells were seeded into a six-well plate with 1 × 105 cells in each well and cultured in an incubator for 24 hr. Then, the cells were infected with LV-shLINC00707, LV-LINC00707, or LV-NC for 48 hr. After that, the cells were digested with trypsin and placed into centrifuge tubes. Three thousand cells in each well were then seeded in 96-well plates. Hundred microliter cell counting kit 8 (CCK8) reagents (Dojindo Molecular Technologies, Tokyo, Japan) were added at 0, 1, 2, 3, and 4 days respectively. After incubation for 4 hr, a microplate reader (Bio-Tek, Winooski, VT) was used.
2.4 5-ethynyl-2′-deoxyuridine incorporation assay
5-ethynyl-2′-deoxyuridine (EdU) proliferation assay (RiboBio, Nanjing, China) was used to test HCC cell proliferation. After infection for 48 hr, the cells were indicated with 50 μM EdU. Apollo staining and DAPI staining were used to detect the EdU positive cells according to the instructions. Incorporation of EdU can identify cells undergoing DNA replication. Image-Pro Plus software (Version 6.0, Media Cybernetics, Bethesda, MD) was used to calculate the percentage of EdU-positive (EdU+) cells.
2.5 Clonogenic assay
A total of 500 cells seeded in six-well plates were indicated in medium containing 10% fetal bovine serum for 2 weeks. Two weeks later, the cells were fixed using methanol and stained using 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO).
2.6 Cell apoptosis analysis
Infected cells were stained using FITC-Annexin V and Propidium iodide (PI). Apoptosis was observed using FACS Calibur (BD Biosciences, San Jose, CA) and analyzed using Flowjo software (Tree Star Corp, San Carlos, CA).
2.7 Cell cycle analysis
The cells were seeded in six-well plates and synchronized at the G1/S boundary. The cells were fixed in 70% ethanol and stained using phosphate-buffered saline containing 50 mg/ml of PI and 20 mg/ml of RNase A.
2.8 Transwell invasion assay
The inserts were coated using a thin layer of Matrigel, and 60 μl Matrigel was diluted by precooled serum-free media in a 1:4 ratio added to the middle of the bottom of the Transwell chamber. The inserts were fixed using 4% methanol and stained with 0.05% crystal violet.
2.9 Quantitative real-time polymerase chain reaction
RNAs were extracted from HCC cells using TRIzol reagent (Invitrogen). RNA reverse transcription was conducted using Prime Script TM RT Master Mix and quantitative polymerase chain reaction (qPCR) was carried out using SYBR Premix Ex Taq II (TaKaRa Bio Technology, Dalian, China). The primers are exhibited in Table 1. Applied Biosystems 7900 Real-Time PCR System (Applied Biosystems, Foster City, CA) was used. The relative expression of target genes was calculated using the 2−ΔΔCt method.
| Genes | Forward (5′~3′) | Reverse(5′~3′) |
|---|---|---|
| GAPDH | TATCGGACGCCTGGTTAC | TATCGGACGCCTGGTTAC |
| LINC00707 | TCACATCTGTGAAAAGAGTGCT | CTGGACTGTGAGTACCAGGC |
- Note. PCR: polymerase chain reaction.
2.10 Western blot
Protein samples were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA). Then, the membranes were incubated with 5% non-fat milk. The protein abundance of GAPDH (1:1,000, Abcam, Cambridge, Britain, cat. no. ab8245) served as a control. Primary antibodies, including anti-p-ERK (cat. no. ab50011), anti-p-AKT (R cat. no. ab8933) anti-p-JNK (cat. no. ab46821), anti-ERK (cat. no. ab54230), anti-AKT (cat. no. ab8805), and anti-JNK (JNK; cat. no. ab201624) at 4°C were used. Next day, the membranes were incubated with a secondary antibody at room temperature for 2 hr. Protein bands were exposed using an EZECL chemiluminescence detection kit for HRP (Millipore).
2.11 Animal studies
Eighteen BALB/c mice (purchased from Shanghai Animal Laboratory Center) were kept in the Experimental Animal Center with appropriate sterile filter-capped cages. The lights were turned on at 7:30 a.m. and turned off at 5:30 p.m. with 22 ± 1°C temperature and 55 ± 5% humidity. Wood shavings, feedstuff, and water were provided in all cages to guarantee environmental enrichment. Six mice were injected in the front dorsum with parental Hep3B cells (5 × 106 each), six were injected with Hep3B infected with LV-shLINC00707, and the other six were injected with Hep3B infected with LV-LINC00707. Two weeks later, tumor sizes were recorded every three days. The length and width of tumor were tested using a caliper, and the volumes were calculated using the following formula: volume (mm3) = length × width × width/2. Six weeks later, the animals were killed by cervical dislocation. This study was strictly according to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
2.12 Immunohistochemistry staining and hematoxylin-eosin staining
Sections were exposed to heat-induced antigen-retrieval in citric acid buffer (pH 7.0), blocked using 5% normal goat serum, and incubated with 3% hydrogen peroxide. The sections were washed, blocked by 10% normal goat serum, and incubated with primary antibodies to Ki-67 (1:500, Abcam). For hematoxylin-eosin (HE) staining, tumor tissues were fixed with 10% formalin. Sections were cut and stained using hematoxylin and eosin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China).
2.13 Statistical analysis
Statistical data analysis was done using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA). Experiments were conducted in triplicate, and data were displayed as mean ± standard deviation (SD). Statistical analysis was carried out using Student's t-test or one-way analysis of variance. Statistical significance was significant when p < 0.05.
3 RESULTS
3.1 LINC00707 was upregulated in HCC cells
First, LINC00707 in HCC cells including Hep3B, HepG2, MHCC97L, SNU449, Huh-7 cells and normal human liver cell line LO2 cells was detected by using real-time PCR. As shown in Figure 1a, LINC00707 was significantly increased in HCC cells compared with LO2 cells. This indicated that LINC00707 might be involved in HCC development.
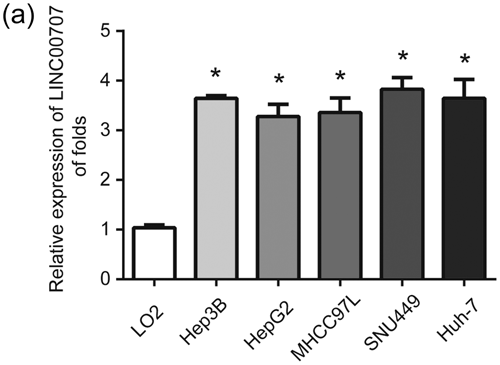
LINC00707 was elevated in HCC cells. (a) LINC00707 expression in Hep3B, HepG2, MHCC97L, SNU449, Huh-7 and LO2 cells. GAPDH was used as a reference control. Three independent experiments were carried out. Error bars stand for the mean ± standard deviation (SD) of at least triplicate experiments. *p < 0.05. HCC: hepatocellular carcinoma
3.2 Knockdown of LINC00707 restrained HCC cell proliferation
To explore whether LINC00707 can affect HCC cell survival, LINC00707 was silenced in Hep3B and SNU449 cells. The cells were infected with LV-NC, LV-shLINC00707, and LV-LINC00707 for 48 hr. As exhibited in Figure 2a, LINC00707 was successfully decreased by LV-shLINC00707 and increased by LV-LINC00707 in HCC cells. Then, we observed that HCC cell survival was greatly repressed after LINC00707 was knocked down in HCC cells (Figure 2b). In addition, EdU assay was also performed, and it was indicated that silencing of LINC00707 could significantly inhibit HCC cell proliferation while overexpression of LINC00707 induced HCC cell proliferation (Figure 2c,d). Next, colony formation assays displayed that HCC cell colony formation capacity was greatly inhibited by LV-shLINC00707 while increased by LV-LINC00707 (Figure 2e). These results implied that overexpression of LINC00707 could promote HCC cell proliferation in vitro.
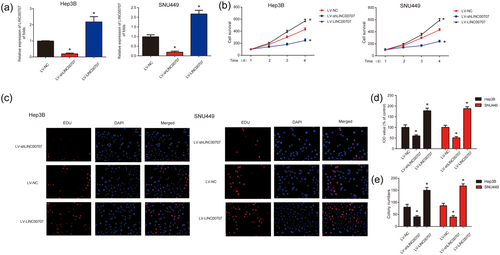
Effects of LINC00707 on HCC cell proliferation. (a) LINC00707 expression in Hep3B and SNU449 cells. Cells were infected with LV-shLINC00707 for 48 hr. (b) Effects of LV-shLINC00707 and LV-LINC00707 on the cell survival of Hep3B and SNU449 cells. CCK8 assay was used to test cell viability. (c,d) Effects of LV-shLINC00707 and LV-LINC00707 on the cell proliferation of Hep3B and SNU449 cells. (e) Effects of LV-shLINC00707 and LV-LINC00707 on the cell proliferation of Hep3B and SNU449 cells. Three independent experiments were carried out. Error bars stand for the mean ± standard deviation (SD) of at least triplicate experiments. *p < 0.05. CCK8: Cell counting kit; HCC: hepatocellular carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
3.3 LINC00707 inhibition induced HCC cell apoptosis and blocked HCC cell cycle progression
Then, flow cytometry was used to detect the effect of LV-shLINC00707 on HCC cell apoptosis and cell cycle. It was observed that apoptosis of HCC cells was dramatically induced by LV-shLINC00707 as shown in Figure 3a,b. Oppositely, upregulation of LINC00707 inhibited HCC apoptosis strongly. In addition, LINC00707 can influence HCC cell cycle progression. As exhibited in Figure 3c–e, LV-shLINC00707 dramatically blocked cell cycle progression in G1 phase while LINC00707 overexpression exhibited a revered effect in Hep3B and SNU449 cells. These results indicated that LINC00707 knockdown induced HCC cell apoptosis and HCC cell cycle arrest in vitro.
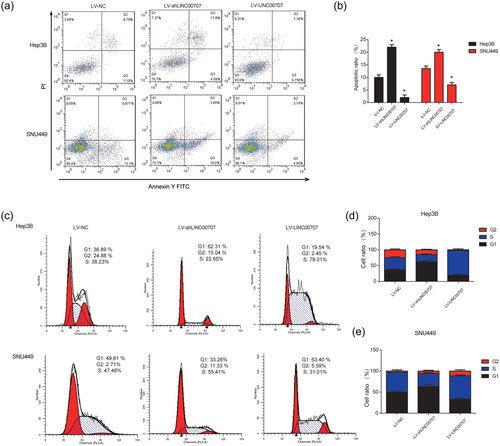
Effects of LINC00707 on HCC cell apoptosis and cell cycle. (a,b) Effects of LV-shLINC00707 and LV-LINC00707 on the cell apoptosis in Hep3B and SNU449 cells. Flow cytometry assay was used to analyze the cell apoptosis. (c,e) Effects of LV-shLINC00707 and LV-LINC00707 on the cell cycle progression. Flow cytometry assay was performed to analyze the cell cycle. Three independent experiments were carried out. Error bars stand for the mean ± standard deviation (SD) of at least triplicate experiments. *p < 0.05. HCC: hepatocellular carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
3.4 LINC00707 downregulation repressed HCC cell invasion
Moreover, to investigate the effect of LINC00707 on HCC invasion, transwell invasion assay was used. As demonstrated in Figure 4a,b, LV-shLINC00707inhibited HCC cell invasion capacity of HCC cells. In addition, LV-LINC00707 promoted HCC cell invasion ability significantly. These results suggested that LINC00707 silence repressed HCC cell invasion.
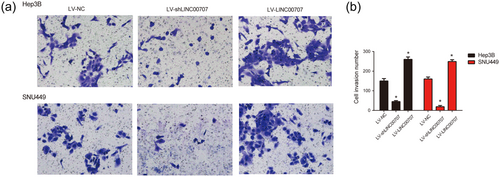
Effects of LINC00707 on HCC cell invasion ability. (a,b) Effects of LV-shLINC00707 and LV-LINC00707 on the cell invasion of Hep3B and SNU449 cells. Transwell migration and invasion assay were conducted to test cell invasive ability. Three independent experiments were carried out. Error bars stand for the mean ± standard deviation (SD) of at least triplicate experiments. *p < 0.05. HCC: hepatocellular carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
3.5 LINC00707 was responsible for the activation of ERK/JNK/AKT in HCC cells
To evaluate the potential molecular mechanism of LINC00707 in HCC cell progression, the ERK/JNK/AKT signaling pathway was investigated. As shown in Figure 5a,b, we observed that decreased expression of LINC00707 inhibited phosphorylation of ERK2, AKT and JNK in Hep3B cells. On the contrary, as shown in Figure 5c,d, LV-LINC00707 induced phosphorylation of ERK2, AKT and JNK. These indicated that LINC00707 was able to mediate the activity of the ERK/JNK/AKT signaling pathway in HCC cells.
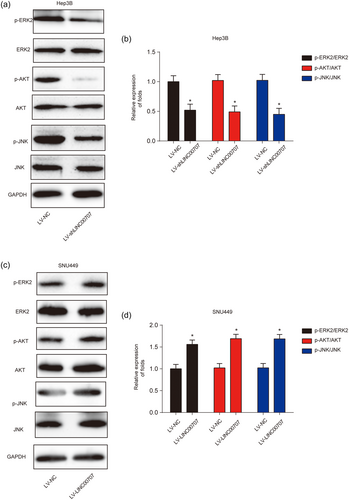
Effects of LINC00707 on ERK/JNK/AKT signaling pathway. (a,b) Effects of LV-shLINC00707 on ERK/JNK/AKT signaling pathway in Hep3B cells. (c,d) Effects of LV-LINC00707 on ERK/JNK/AKT signaling pathway in SNU449 cells. Three independent experiments were carried out. Error bars stand for the mean ± standard deviation (SD) of at least triplicate experiments. *p < 0.05 [Color figure can be viewed at wileyonlinelibrary.com]
3.6 LINC00707 modulated HCC development through activating ERK/JNK/AKT signaling pathway in vivo
Furthermore, to explore whether LINC00707 could regulate HCC development via the ERK/JNK/AKT signaling pathway in vivo, a Hep3B cell nude mouse xenograft model was used to investigate whether LINC00707 can regulate HCC development in vivo. Six mice were injected with Hep3B cells infected with LV-NC, six were injected with Hep3B cells infected with LV-shLINC00707, while the other six were injected with Hep3B cells infected with LV-LINC00707. Tumors were peeled and manifested in Figure 6a. LV-shLINC00707 inhibited tumor volume and LV-LINC00707 increased tumor growth in a time-dependent manner (Figure 6b). HE staining results were shown, and the immunohistochemistry (IHC) data demonstrated that Ki-67 was greatly repressed by LV-shLINC00707 while enhanced by LV-LINC00707 (Figure 6c). Meanwhile, LINC00707 knockdown inhibited ERK/JNK/AKT signaling in vivo whereas LINC00707 overexpression activated the ERK/JNK/AKT pathway as shown in Figure 6d. These data revealed that LINC00707 knockdown inhibited HCC development through targeting ERK/JNK/AKT signaling in vivo.
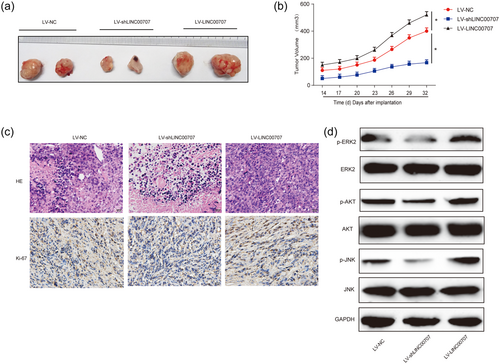
Effects of LINC00707 on HCC progression in vivo. Eighteen 8-week old female BALB/c nude mice were injected with Hep3B cells infected with LV-NC (six mice), LV-shLINC00707 (six mice) or LV-LINC00707 infected Hep3B cells (six mice). (a) Solid tumors were peeled from mouse subcutaneous tissue. (b) Tumor volume in a time-dependent course. (c) H&E staining and IHC staining of Ki-67 in tumor tissues. (d) Protein expression of p-ERK2, p-AKT and p-JNK in the tumor tissues from the mice. Three independent experiments were carried out. Error bars stand for the mean ± standard deviation (SD) of at least triplicate experiments. *p < 0.05. HCC: hepatocellular carcinoma; H&E: hematoxylin-eosin; IHC: immunohistochemistry [Color figure can be viewed at wileyonlinelibrary.com]
4 DISCUSSION
Recently, lncRNAs have been recognized as important biomarkers in various cancers, including HCC. Hence, it is urgent to identify prognostic lncRNAs for HCC diagnosis and treatment. In our current study, we found that LINC00707 was significantly upregulated in HCC cells. Inhibition of LINC00707 was able to depress HCC cell proliferation, cell colony formation and induced cell apoptosis, while overexpression demonstrated a reversed effect. In addition, knockdown of LINC00707 restrained HCC cell cycle progression in G1 phase, meanwhile, cell invasion ability was also inhibited by LV-shLINC00707. Here, we observed that LINC00707 could promote HCC progression through activating ERK/JNK/AKTsignaling.
Recent studies have indicated that multiple lncRNAs can drive forces of carcinogenesis. For example, LINC00210 activates Wnt/β-catenin signaling in HCC dependent on CTNNBIP1 (X. Fu et al., 2018). LINC00673 can promote HCC progression through negatively regulating miR-205 (L.G. Zhang, Zhou, Zhou, Lv, & Li, 2017). LncRNA00364 can repress HCC cell proliferation via regulating p-STAT3-IFIT2 signaling (Tang et al., 2017). In our study, we focused on the biological role of LINC00707, a 3,087 bp non-coding RNA, located on chromosome 10p14. Previously, it has been reported that LINC00707 can promote lung cancer progression by regulating Cdc42 (Ma et al., 2018). LINC00707 is increased in the early-stage group compared with the advanced-stage group of melanoma patients (Yang, Xu, & Zeng, 2018). Here, we found that LINC00707 was increased in HCC cells and silencing of LINC00707 significantly inhibited HCC progression in vitro. In our future study, if it is possible, we will collect HCC samples to test LINC00707 expression.
It is well accepted that the abnormal molecular signaling pathways are involved in the biological process of HCC (Bruix, Sherman, & American Association for the Study of Liver D, 2011; Galuppo, Ramaiah, Ponte, & Gedaly, 2014; Q. Zhou, Lui, & Yeo, 2011). The MAPK signaling pathway is the key pathway that can participate in cell proliferation, apoptosis, and differentiation (Wagner & Nebreda, 2009). ERK is conserved, and its abnormal expression is closely related with human diseases (Thum et al., 2008; Whelan, Hollis, Cha, Asch, & Lee, 2012). For example, lncRNA BANCR can promote endometrial cancer cell progression ERK Signaling Pathway (D. Wang, Wang, Wang, Long, & Ren, 2016). MALAT1 can promote gallbladder cancer development through activating ERK and AKT (Wu et al., 2014). LncRNA URHC can regulate HCC cell progression through ERK/MAPK (W.H. Xu et al., 2014). Downregulation of BANCR suppresses HCC development through inactivating ERK/MAPK (Li et al., 2017). MAPK signaling comprises a family of protein kinases (JNK and ERK), which play important roles in cellular differentiation, proliferation, and survival (Boutros, Chevet, & Metrakos, 2008). The JNK pathway has been reported to regulate various physiological processes, including cell differentiation, cell proliferation, cell death, and cell survival (Kumar et al., 2015). In addition, AKT is a serine/threonine kinase, and it is a key player in cell signals that are important for cell death and survival (Song, Ouyang, & Bao, 2005). In our study, we observed that knockdown of LINC00707 inhibited HCC development via inactivating ERK/JNK/AKTsignaling while overexpression activated the ERK/JNK/AKT pathway.
In conclusion, we indicated that LINC00707 exhibited an oncogenic role in HCC development. It was found that silencing of LINC00707 blocked HCC progression through inactivating the ERK/JNK/AKT signaling pathway. Taken together, our results implied that LINC00707 was involved in HCC progression and it might act as a novel biomarker for HCC.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
FUNDING
This study was supported by the Guangxi Clinic Medicine Research Center of Hepatobiliary Diseases (AD17129025).