Upregulation of long noncoding RNA XIST is associated with poor prognosis in human cancers
Abstract
Growing evidence from recent studies has shown that the X-inactive specific transcript (XIST), a well-known long noncoding RNA involved in early embryonic development, is aberrantly regulated in various human cancers. However, the prognostic value of XIST in cancers remains uncharacterized. In this study, we searched PubMed, Web of Science, and Embase to collect all relevant studies, and a meta-analysis was performed to explore the association of XIST expression with overall survival (OS) and clinicopathological parameters. We demonstrated that high XIST expression was associated with poor OS (hazard ratio = 1.76; 95% confidence intervals [CI], 1.56–1.98; p < 0.001). In addition, increased XIST expression was found to be associated with lymph node metastasis (odds ratio [OR] = 2.06; 95% CI, 1.46–1.90; p < 0.001), distant metastasis (OR = 2.93; 95% CI, 2.00–4.28; p < 0.001), tumor size (OR = 2.66; 95% CI, 1.86–3.81; p < 0.001), poor differentiation (OR = 1.45; 95% CI, 1.00–2.10; p = 0.049), and advanced tumor stage (OR = 3.35; 95% CI, 2.25–5.00; p < 0.001), but not with age (OR = 0.82; 95% CI, 0.59–1.15; p = 0.251) or gender (OR = 0.92; 95% CI, 0.70–1.19; p = 0.512). Our meta-analysis showed that XIST may be a useful common biomarker for predicting prognosis in patients with cancer.
1 INTRODUCTION
In eutherian mammals, X chromosome inactivation (XCI) transcriptionally silences one X chromosome in the female to balance gene expression between the sexes (Lyon, 1961; Wutz, 2011). XCI is triggered by the expression of the X-inactive specific transcript (XIST), a long noncoding RNA that binds chromosome from which it is transcribed (Brockdorff et al., 1991; Brown et al., 1991). XIST tethers silencing factors in cis, which modify the chromatin to become a heterochromatic state (Jeon & Lee, 2011). XCI takes place only once during early embryonic development. After XCI initiation, the inactive X chromosome enters the maintenance phase where XIST is dispensable (Jonkers et al., 2008).
In recent years, XIST has attracted increasing interest because aberrant expression of XIST is reported in a series of human cancers. In colorectal cancer, the expression of XIST was significantly upregulated in tumor tissues compared with that of adjacent normal tissues and upregulated XIST expression is associated with poor prognosis (Chen et al., 2017; H. Song et al., 2017). In addition, upregulated XIST expression was observed in glioma (Du et al., 2017), gastric cancer (Chen et al., 2016; L. Ma et al., 2017), pancreatic cancer (Sun, Zhang, & Cui, 2018; Wei, Liu, Lu, Yang, & Tang, 2017), non-small cell lung cancer (Fang, Sun, & Gong, 2016), nasopharyngeal carcinoma (P. Song, Ye, Zhang, Peng, & Zhou, 2016), esophageal squamous cell carcinoma (Wu et al., 2017), and bladder cancer (Hu et al., 2017), leading to poor clinicopathologic features and short survival time. In contrast, two studies demonstrated downregulated XIST expression (Chang, Chen, Wang, Wu, & Sun, 2017; W. Ma et al., 2017), whereas another study revealed upregulated XIST expression in hepatocellular carcinoma (Kong et al., 2018). Moreover, studies reported opposite results in cervical squamous cell carcinoma (Kobayashi et al., 2016; Zhu, Zheng, Yu, Zhou, & Wang, 2018) and in osteosarcoma (Li, Wu, Li, & Li, 1961; Yang, Wu, Wang, & Wei, 2018; Zhang & Xia, 2017). Therefore, the prognostic value of XIST for patients with cancer is still unclear.
Regardless of the inconsistent results, XIST is still a potent biomarker for predicting cancer prognosis. The meta-analysis methodology provides a necessary means to increase statistical power and to resolve inconsistencies or discrepancies among different studies. In the present study, we performed a meta-analysis of eligible studies to obtain an evidence-based result on the prognostic role of XIST in various cancers.
2 METHODS
2.1 Literature search
A literature search was conducted on the online databases PubMed, Web of Science, and Embase (up to August 1, 2018). The keywords were (“XIST” OR “X-inactive specific transcript”) AND (“prognostic” AND “prognosis” OR “survival”).
2.2 Study selection
The inclusion criteria of this meta-analysis were (a) research on the association between XIST and cancer prognosis, (b) division of patients into two groups: high and low XIST expression, and (c) sufficient data for the computation of odds ratio (OR) or hazard ratio (HR) with 95% confidence intervals (CI). The exclusion criteria for our meta-analysis were (a) articles that are case reports, editorials, and reviews, (b) non-English language and non-human studies, and (c) studies without available data.
2.3 Quality assessment for studies
Study quality was assessed in accordance with Newcastle-Ottawa quality assessment scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The NOS score items, including the selection of cohorts, comparability of cohorts, and assessment of outcome, were extracted. The total scores ranged from 0 to 9. The studies were considered of high quality if their scores were higher or equal to 6.
2.4 Data extraction
The following information from each eligible study was extracted: (a) publication details, including first author, year of publication, study region, sample size, cancer type, and detection method; (b) clinicopathological data, including age, gender, lymph node metastasis (LNM), distant metastasis (DM), tumor size, differentiation and tumor stage, and (c) HR and 95% CI for overall survival (OS) and disease-free survival. If a study reported the results of univariate and multivariate analysis at the same time, only the latter was extracted. If only the Kaplan–Meier curve was provided in the article, the survival data were extracted from the graphical curve using Engauge Digitizer v10.4 software (http://markummitchell.github.io/engauge-digitizer/). HR and 95% CI were then determined using the published method (Tierney, Stewart, Ghersi, Burdett, & Sydes, 2007).
2.5 Statistical analysis
Statistical analysis was performed by using Stata SE14.0 software (StataCorp, Texas). XIST expression was categorized into the high-expression group and the low-expression group according to the original published articles. OR with 95% CI were combined to evaluate the association between XIST expression and the clinicopathological features. Prognostic role of XIST expression on OS was assessed by estimation of the pooled HR and matching 95% CI. Statistical heterogeneity between studies was analyzed by using the I2 statistics and chi-square-based Q test. When there was no significant heterogeneity (I2 < 50% and p > 0.05), the fixed-effect model was chosen; otherwise, the random-effect model was adopted to enhance the stability of the meta-analysis. The sensitivity analysis was conducted to investigate the robustness of the overall meta-analysis results. The suspected factors for heterogeneity were investigated using metaregression models. Publication bias was appraised by Begg’s test and Egger’s test, respectively. All p values were calculated with a two-tailed test, and p < 0.05 was considered as statistically significant.
3 RESULTS
3.1 Characteristics of eligible studies
Our initial search retrieved 135 potentially relevant papers from PubMed, Web of Science, and Embase. Ultimately, 18 studies met the eligibility criteria and were included in the meta-analysis (Chen et al., 2016, 2017; Du et al., 2017; Fang et al., 2016; Hu et al., 2017; Kobayashi et al., 2016; Kong et al., 2018; Li, Wu, Li, & Li, 2017; L. Ma et al., 2017; W. Ma et al., 2017; P. Song et al., 2016, H. Song et al., 2017; Sun et al., 2018; Wei et al., 2017; Wu et al., 2017; Yang et al., 2018; Zhang & Xia, 2017; Zhu et al., 2018). A flow diagram of the study selection process is shown in Figure 1. In total, 1,428 patients were included in this study. The publication time of these studies ranged from 2016 to 2018. Fourteen studies came from China and one came from Japan. The 18 studies covered 11 different types of cancer, including colorectal cancer (n = 2), glioma (n = 1), gastric cancer (n = 2), osteosarcoma (n = 3), hepatocellular carcinoma (n = 2), pancreatic cancer (n = 2), cervical squamous cell carcinoma (n = 2), non-small cell lung cancer (n = 1), nasopharyngeal carcinoma (n = 1), esophageal squamous cell carcinoma (n = 1), and bladder cancer (n = 1). In all of the studies, XIST expression was detected by quantitative reverse transcription-polymerase chain reaction. The main features of the 18 studies used for the present meta-analysis are summarized in Table 1.

Flow diagram of the study search and selection process
| Study | Country | Sample size | Cancer | Method | Outcome |
|---|---|---|---|---|---|
| Chen et al. (2017) | China | 115 | CRC | qRT-PCR | OS |
| H. Song et al. (2017) | China | 50 | CRC | qRT-PCR | DFS |
| Du et al. (2017) | China | 69 | Glioma | qRT-PCR | OS |
| L. Ma et al. (2017) | China | 98 | GC | qRT-PCR | OS |
| Chen et al. (2016) | China | 106 | GC | qRT-PCR | OS |
| Zhang & Xia (2017) | China | 41 | Osteosarcoma | qRT-PCR | OS |
| Li et al. (2017) | China | 145 | Osteosarcoma | qRT-PCR | OS |
| W. Ma et al. (2017) | China | 68 | HCC | qRT-PCR | OS |
| Wei et al. (2017) | China | 64 | PC | qRT-PCR | OS |
| Kobayashi et al. (2016) | Japan | 49 | CSCC | qRT-PCR | OS |
| Fang et al. (2016) | China | 53 | NSCLC | qRT-PCR | OS |
| P. Song et al. (2016) | China | 108 | NPC | qRT-PCR | OS |
| Wu et al. (2017) | China | 127 | ESCC | qRT-PCR | OS |
| Hu et al. (2017) | China | 52 | BLC | qRT-PCR | OS |
| Kong et al. (2018) | China | 52 | HCC | qRT-PCR | OS |
| Sun et al. (2018) | China | 139 | PC | qRT-PCR | OS |
| Yang et al. (2018) | China | 40 | Osteosarcoma | qRT-PCR | OS |
| Zhu et al. (2018) | China | 52 | CSCC | qRT-PCR | OS |
- Note. BLC: bladder cancer; CRC: colorectal cancer; CSCC: cervical squamous cell carcinoma; DFS: disease-free survival; ESCC: esophageal squamous cell carcinoma; GC: gastric cancer; HCC: hepatocellular carcinoma; NPC: nasopharyngeal carcinoma; NSCLC: non-small cell lung cancer; OS: overall survival; PC: pancreatic cancer; qRT-PCR: quantitative reverse transcription-polymerase chain reaction.
3.2 Relationship between XIST expression and clinicopathological parameters
There are 7 and 12 studies describing cancer patients of age and gender, respectively. Pooled analysis showed that XIST expression did not correlate significantly with age (Figure 2a) or gender (Figure 2b). A total of eight studies evaluated LNM according to XIST expression. There was significant heterogeneity among these studies (I2 = 55.9%; p = 0.026). Subsequently, a random-effect model was adopted to analyze the data. The OR of LNM for the high XIST -expression group versus the low expression group was 2.06 (95% CI, 1.46–2.90; p < 0.001; Figure 2c). A total of six studies evaluated DM according to XIST expression. The heterogeneity among studies was not statistically significant (p = 0.842; I2 = 0.0%). The fixed-effect model was applied to calculate the pooled OR 2.93 (95% CI, 2.00–4.28; p < 0.001; Figure 2d). These aggregated results suggested that cancer patients with high XIST expression were more susceptible to LNM and DM than patients with low XIST expression. In addition, we found that high XIST expression was positively associated with tumor size (OR = 2.66; 95% CI, 1.86–3.81; p < 0.001; Figure 2e), poor tumor differentiation (OR = 1.45; 95% CI, 1.00–2.10; p = 0.049; Figure 2f), and advanced clinical stage (OR = 3.35; 95% CI, 2.25–5.00; p < 0.001; Figure 2g). Because of insufficient data, we were unable to determine the association between high XIST expression and other clinicopathological parameters.
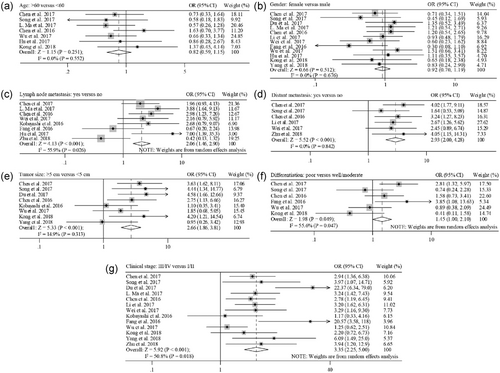
Meta-analysis for the relationship between XIST expression and clinicopathological parameters. The investigated clinicopathological parameters are: (a) age, (b) gender, (c) lymph node metastasis, (d) distant metastasis, (e) tumor size, (f) differentiation, and (g) clinical stage. XIST: X-inactive specific transcript
3.3 Relationship between XIST expression and OS
The relationship between XIST expression and OS was evaluated in 13 studies including 1,428 patients. Because significant heterogeneity was observed among these studies (I2 = 95.8%; p < 0.001), a random-effect model was used to pool the results. The pooled HR indicated that XIST expression was negatively associated with OS (HR = 1.76; 95% CI, 1.56–1.98; p < 0.001; Figure 3a). Sensitivity analysis was performed to assess the effect of individual study. By successively omitting each individual study, the result was not significantly affected, suggesting that the result of the meta-analysis was robust (Figure 3b). Subsequent subgroup analysis stratified by cancer types revealed that a significantly unfavorable OS was observed in patients with digestive tract cancers (HR = 2.94; 95% CI, 2.28–3.80) and, to a lesser extent, in patients with nondigestive tract cancers (HR = 1.60; 95% CI, 1.41–1.82) (Figure 4a). Furthermore, a metaregression was conducted to explore the potential factors responsible for the heterogeneity. We found that the tumor type (digestive tract cancers vs nondigestive tract cancers) was a significant factor for the heterogeneity of the meta-analysis results (p = 0.001; Figure 4b). High heterogeneity was still observed in nondigestive tract cancers (I2 = 96.4%; p < 0.001), but not in digestive tract cancers (I2 = 0.0%; p = 0.594; Figure 4a).

Meta-analysis for the relationship between XIST expression and overall survival. (a) Forest plot for the association between XIST expression and overall survival. (b) Sensitivity analysis of individual study effect on the pooled HR. XIST: X-inactive specific transcript; HR: hazard ratio
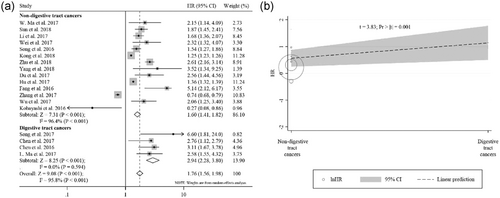
Subgroup analysis of overall survival by cancer types. (a) Forest plot for the association between XIST expression and overall survival stratified by cancer types. (b) Metaregression analysis for assessment of heterogeneity. XIST: X-inactive specific transcript
3.4 Analysis of publication bias
Finally, Begg’s funnel plot (Figure 5a) and the Egger’s linear regression test (Figure 5b) were used to assess publication bias. The tests revealed p values greater than 0.05 (Begg’s p = 0.426 and Egger’s p = 0.101). Thus, there was no publication bias in this meta-analysis.
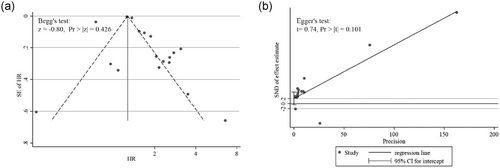
Assessing publication bias in the meta-analysis of overall survival. (a) Begg’s test and funnel plot with pseudo 95% CI. (b) Egger’s test and linear regression plot. CI: confidence intervals
4 DISCUSSION
XIST was first long noncoding RNA transcript to be discovered in mammals (Brockdorff et al., 1991; Brown et al., 1991). After more than two decades of extensive research, XIST-mediated initiation and spreading of XCI in female somatic cells is well established (da Rocha & Heard, 2017). Recently, XIST dysregulation has been found in several cancers, suggesting that XIST could have a potential diagnostic power in cancer. Here, we performed a meta-analysis of 18 studies, including 1,428 patients, who were used to evaluate the prognostic value of XIST and the relationship between increased XIST expression and clinicopathological characteristics in different cancers. The pooled results demonstrated that increased XIST expression was significantly and positively associated with LNM, DM, tumor size, poor differentiation, and advanced clinical stage, but not with age or gender. Meanwhile, we found that OS was lower for patients with high XIST expression than for those with low expression. Sensitivity analysis indicated that no single study impacted the overall results, suggesting that the pooled result was robust. Furthermore, no obvious publication bias was observed among our included studies. Therefore, XIST may be a useful biomarker for predicting prognosis in patients with cancer.
Several studies have elucidated the molecular mechanisms underlying the association between elevated XIST expression and poor outcomes of patients with cancer. It has been suggested that XIST may suppress the functions of microRNAs by acting as a competitive endogenous RNA (ceRNA). Chen et al. (2017) found that XIST regulates colorectal cancer progression and metastasis by competing for miR-200b-3p to modulate the expression of zinc finger E-box-binding homeobox 1 (ZEB1), whereas H. Song et al. (2017) showed that XIST promoted colorectal cancer cell proliferation by antagonizing the inhibitory effect of miR-132-3p on mitogen-activated protein kinase 1 (MAPK1). XIST and miR-29c reciprocally inhibited each other in glioma (Du et al., 2017). In gastric cancer, it was reported that XIST regulates cancer cell proliferation and invasion via miR-101/enhancer of zeste homolog 2 (EZH2) axis (Chen et al., 2016) and miR-497/metastasis-associated in colon cancer 1 axis (L. Ma et al., 2017). Notably, XIST also promotes esophageal squamous cancer through the miR-101/EZH2 axis (Wu et al., 2017). Wei et al. (2017) revealed that XIST contributes to pancreatic cancer proliferation via miR-133a/epidermal growth factor receptor. P. Song et al. (2016) demonstrated that XIST functioned as the ceRNA of miR-34a-5p to regulate transcription factor E2F3 in nasopharyngeal carcinoma. Apart from a role of ceRNA, XIST could directly interact with EZH2 to suppress transcription of its target gene Krüppel-like factor 2 (KLF2) in non-small cell lung cancer (Fang et al., 2016). These results suggest that XIST plays a key role in cancer progression with different mechanisms in various cancer subtypes.
Notably, there are discordant results in hepatocellular carcinoma, cervical squamous cell carcinoma, and osteosarcoma. In hepatocellular carcinoma, the tumor suppressor activity of XIST was exerted by upregulating phosphatase and tensin homolog deleted on chromosome ten (PTEN) expression via miR-181a (Chang et al., 2017), whereas the oncogenic role of XIST was exerted by upregulated MAPK1 expression via miR-194-5p (Kong et al., 2018). The underlying mechanism of XIST as a tumor suppressor in cervical squamous cell carcinoma is unexplored (Kobayashi et al., 2016); however, its oncogenic role in cervical squamous cell carcinoma was reported to attribute to the upregulation of RNA binding protein FUS through miR-200a (Zhu et al., 2018). XIST upregulated programmed cell death protein 4 (PDCD4) expression by interacting with miR-21-5p and inhibited osteosarcoma cell growth and metastasis (Zhang & Xia, 2017). Meanwhile, it was demonstrated that XIST promoted osteosarcoma progression by targeting yes-associated protein 1 (YAP) via miR-195-5p (Yang et al., 2018). These contradictory results deserve further investigation.
There are some limitations that should be noted for the current study. First, there was no consensus on the cutoff value for defining high and low XIST expression. Second, most of the included patients with cancer were from China. Thus, more clinical data from other ethnic groups are needed to confirm our results. Third, in some cases, HR and 95% CI were extracted from the Kaplan–Meier curves, which are less reliable than those directly obtained from the original studies. Finally, most studies tended to report positive results rather than negative results. Therefore, it is possible that our meta-analysis results might exaggerate the prognostic value of elevated XIST expression on survival and clinicopathological characteristics in cancers.
In conclusion, our meta-analysis suggests that the upregulation of XIST is a risk factor for poor clinical outcomes in cancers. For future clinical application, well-designed studies related to specific cancer types with large sample sizes are needed to confirm the prognostic value of XIST in various cancers.
ACKNOWLEDGMENTS
This work was funded by National Natural Science Foundation of China (grant number 31771665 to Ji-Long Liu). The funder has no role in study design, data collection, analysis, or interpretation.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest that could be perceived as prejudicing the impartiality of the research reported.