microRNA-874 inhibition targeting STAT3 protects the heart from ischemia–reperfusion injury by attenuating cardiomyocyte apoptosis in a mouse model
Abstract
MicroRNAs (miRs) were involved in numerous cardiovascular diseases, especially ischemic heart diseases, but the miR changes during cardiac ischemia–reperfusion (I/R) injury following sevoflurane (SEV) preconditioning are still unknown. This study aims to investigate the effect of miR-874 on cardiac I/R injury in mouse models pretreated with SEV. Following establishment of mouse models with myocardial I/R injury, mice were pretreated with SEV. The functional mechanism of miR-874 in I/R injury was explored when miR-874 and the Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling pathway were inhibited. Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP biotin nick-end labeling (TUNEL) staining was used to detect cardiomyocyte apoptosis and dual luciferase reporter gene assay to identify the targeting relationship between miR-874 and STAT3. Expression of the JAK2/STAT3 signaling pathway and apoptosis-related genes was determined. Initially, upregulated miR-874 was observed in I/R mice. Then, miR-874 inhibition improved cardiac function of I/R mice, inhibited cardiomyocyte apoptosis (also shown as decreased Bcl-2 associated X protein B [Bax] and increased B-cell lymphoma-2 [Bcl-2]), and activated the JAK2/STAT3 signaling pathway. STAT3, a target gene of miR-874, was upregulated following miR-874 inhibition. Finally, we also observed that the effect of miR-874 was lost when the JAK2/STAT3 signaling pathway was blocked. The findings indicate miR-874 as a contributory role in cardiac I/R injury, with miR-874 inhibition alleviating cardiac I/R injury in mice following SEV pretreatment by targeting STAT3 through the JAK2/STAT3 signaling pathway.
1 INTRODUCTION
Ischemic heart disease is the main cause of death in cardiovascular diseases, and the blood supply restoration, such as reperfusion therapy, is believed to be the most effective treatment of cardiac ischemia (L. Yang et al., 2016). However, during reperfusion of ischemic myocardium with oxygen and substrate-rich blood, it may cause or even exacerbate irreversible damage and deteriorate heart function, which is ischemia–reperfusion (I/R) injury (Zhang & Ren, 2014). Anesthetics like sevoflurane (SEV) and isoflurane are reported to be valid therapeutic drugs for I/R injury (Yao, Fang, Shi, & Li, 2010). Besides its fast induction, rapid recovery, and low possibility of “coronary arterial steal,” SEV is proved to protect the heart against myocardial I/R injury, enhancing recovery of cardiac function, reducing myocardial enzyme release, and lowering infarct size when utilized as pretreatment or posttreatment agents (Gong, Yao, Fang, & Li, 2012). However, the mechanism of SEV mediating cardioprotection is still unknown (Zhao et al., 2013).
As a series of endogenous 18–22-nucleotide RNAs, microRNAs (miRs) played an important regulatory role in animals and plants through targeting messenger RNAs (mRNAs) for the division or translation repression (Wang et al., 2016). Moreover, the abnormal expression of miRs plays an important part in the development and progression of tumors and miR-874, one of the miRNAs, targets oncogene AQP3 in gastric cancer modulating cell migration, growth, and invasion (B. Jiang et al., 2014). Inhibited miR-135b-5p is reported to prevent myocardial I/R injury through activating the Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling pathway via mediating anesthesia with SEV inhalation (X. J. Xie et al., 2017). The JAK/STAT signaling pathway, which comprises receptor-associated cytosolic tyrosine kinases (JAKs) family that phosphorylate a tyrosine residue in cognate of STATs, is thought to play an obligatory role in various physiological processes like development and differentiation of cells and related to myocardial I/R injury (Jia et al., 2015). STAT3 as an antiapoptotic factor, plays an important role in the regulation of gene expression and mitochondrial electron transport during cell stress; inactivation of electron flow by STAT3-dependent manner contributes to lower ROS production in the mitochondria and reduced release of cytochrome c during ischemia and probably other forms of cellular stress (Szczepanek et al., 2011). Moreover, recruitment of STAT3 is important in upregulation of cardiac protective and antiapoptotic proteins by suppressing the death receptor pathway and mitochondrial pathway (Bolli et al., 2011). There are few studies covering the role of miR-874 in myocardial I/R injury concerning the JAK2/STAT3 signaling pathway. In this study, therefore, we investigated the potential effect of miR-874 on myocardial I/R injury in mice models pretreated with SEV, and the mechanism concerning the JAK2/STAT3 signaling pathway.
2 MATERIALS AND METHODS
2.1 Ethics statement
All the animal experiments and the animal welfare were in accordance with the Guide for the Care and Use of Laboratory Animal by International Committees.
2.2 Animal model establishment
Forty-eight healthy male Wistar mice with close age, weighing 18–22 g, were provided by Laboratory Animal Center of People’s Liberation Army Academy of Military Medical Sciences (Beijing, China). Mouse models of myocardial I/R injury (I/R mice) were established in randomly selected 36 mice. The mice were anesthetized by intraperitoneal injection of 2% pentobarbital sodium (WS20051129, 50 mg/ml; Sinopharm Group Chemical Reagent Co., Ltd., Shanghai, China). Electrocardiogram (ECG) was connected to mouse limbs to monitor electrode. The mice were artificially ventilated with animal ventilator, with the respiratory rate at 60 times/min, indoor air as air supply and the tidal volume of 13–15 ml. The left fourth intercostal spaces were opened and 6-0 noninvasive absorbable sutures were used to pass through the left anterior descending coronary artery and to block the vessels. Coronary artery occlusion was successfully made when ECG monitor showed significant elevation of ST segment. After 30 min, the suture was loosened, allowing the vessel to pass again. When ECG monitor showed ST segment decrease, the models of myocardial I/R injury were successfully established.
Random number table method was used to randomly divide the mice into seven groups. (a) In the control group, six mice did not receive any treatment. (b) In the sham group, six mice did not receive model establishment but were sutured with 6-0 noninvasive absorbable sutures after thoracotomy to pass through the left anterior descending coronary artery without blocking the vessels. (c) In the I/R group, six I/R mice were randomly selected without any treatment. (d) In the SEV group, six I/R mice were randomly selected. After ischemia for 30 min, the mice inhaled 2.8% SEV (5 × 141; Abbott, Chicago, IL) for 2 min before reperfusion, continued for 5 min, followed by reperfusion for 120 min. (e) In the SEV + miR-874 inhibitor group, six mice were randomly selected from I/R mice. About 24 hr before I/R injury, miR-874 inhibitor (0.2 μl/g; GenePharma Co., Ltd., Shanghai, China) was injected into myocardium of the mice and chest was closed. Then the mice were treated the same as that in the SEV group. (f) In the SEV + AG490 group, six mice were randomly selected from I/R mice and injected intravenously with AG490 (3 μg/g; JAK2/STAT3 inhibitor; HY-12000; Sigma-Aldrich Chemical Company, St. Louis, MO) 5 min before reperfusion. Then the mice were treated the same as that in the SEV group. (g) In the SEV + miR-874 inhibitor + AG490 group, six mice were selected from I/R mice randomly, injected intramyocardially with miR-874 inhibitor (0.2 μl/g) 24 hr before I/R injury and injected intravenously with AG490 (3 μg/g). Then the mice were treated the same as that in the SEV group.
2.3 Transthoracic echocardiography
The mice in each group were anesthetized by intraperitoneal injection of 2% pentobarbital sodium. Transthoracic echocardiography was conducted at the time point of 30 min before ischemia (T0), 30 min after ischemia (T1), 120 min after reperfusion (T2), 4 hr after reperfusion (T3), 8 hr after reperfusion (T4), and 12 hr after reperfusion (T5). Acuson Sequoia 512 high frequency ultrasonic diagnostic equipment (Acuson, Malvern, PA) was used, with a probe frequency of 8.5 MHz and a scan speed of 100 mm/s. The mice were anesthetized via injection of 2% pentobarbital sodium and fixed on an experiment table. The left papillary muscles and left ventricular long axis section were taken to measure the M curve. The mean left ventricular end systolic diameter (LVSD, mm), left ventricular end diastolic diameter (LVDD, mm), left ventricular end systolic volume (LVSV), and left ventricular end diastolic volume (LVDV) were calculated by continuously measuring three cardiac cycles. Left ventricular ejection fraction (LVEF) and left ventricular shortening fraction (LVFS) were obtained by Simpson. The calculation formulas were: LVEF = (LVDV − LVSV)/LVDV × 100% and LVFS = (LVDD − LVSD)/LVDD × 100%. The heart rate (HR) and mean arterial pressure (MAP) were measured at the same time. These heart function parameters were used to evaluate cardiac function in mice. The experiment was repeated three times and its mean value was collected.
2.4 Hematoxylin–eosin (HE) staining
At the 24th hr after reperfusion, mice were killed. The mouse myocardial tissues were fixed in 4% formaldehyde for 6 hr, embedded in paraffin, cut into 3-μm sections, and baked at 60°C overnight. The sections were dewaxed with xylene I and xylene II for 20 min, respectively, dehydrated in graded ethanol (100%, 100%, 95%, 80%, and 70%, each for 5 min, respectively), and stained for 10 min with hematoxylin. Followed by coloration to blue by running water for 15 min, the sections were stained for 30 s with eosin, washed with distilled water to remove the red staining, dehydrated with alcohol, cleared by xylene, and then mounted with neutral gum. Histopathological examination was performed and photographed, and the myocardial tissue coloration, distribution, and the staining intensity were observed. Pathological changes such as necrosis and edema were examined and random images in each group were collected using the morphological image analysis system at 200 times magnification. The experiment was repeated three times in each group. The Billingham score was used to evaluate myocardium damage. According to the percentage of cardiac myocytes in vacuolation and/or myofibrillar deficiency: 0 point was regarded as no cell injury, 1 point as ≤5%, 1.5 points as 6%–15%, 2 points as 16%–25%, 2.5 points as 26%–35%, and 3 points as ≥36%. Ten visual fields were randomly selected from each section and the average value was obtained as the myocardium damage score (Dziegiel, Surowiak, & Zabel, 2002).
2.5 Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP biotin nick-end labeling (TUNEL) staining
At the 24th hr after reperfusion, mice were killed. The myocardial tissues of mice were fixed in 4% paraformaldehyde, dehydrated conventionally, embedded in paraffin, cut into 3-μm slices, attached on a polylysine adhesion slide, and baked at 65°C for 1 hr. Then the slices were dewaxed to water. The proper amount of water was added into the rice cooker (water depth is 3 cm away from the plastic dyeing vat) and boiled. The slices were placed in a plastic dyeing rack and then placed in a vat containing 0.01 mol/L citric acid solution (pH 6.0). The vat was put in the rice cooker, boiled for 10 min, and kept warm for 20 min after power off. Finally, the dye cylinder was removed, cooled at room temperature, washed with phosphate buffer, and then marked by DNA fragment. The remaining operation was conducted according to TUNEL Kit (Bao Ling Man Science and Technology Co., Ltd., Beijing, China) instructions. The experimental results were expressed by the number of cells with apoptotic nuclei and the total number of cells under six random high-magnification fields of vision per slice. The apoptosis index (AI) was the number of apoptotic nuclei in 100 nuclei. AI was calculated as follows: AI = TUNEL positive cells/total cell number × 100%. The experiment was repeated three times in each group and its mean value was taken.
2.6 Dual luciferase reporter gene assay
The target gene of miR-874 was analyzed at a biological prediction website miRNA.org and corresponding sequences containing the target sites were obtained. The DNA of the cardiomyocytes was extracted according to the protocols of TIANamp Genomic DNA Kit (TIANGEN Biotechnology Co., Ltd., Beijing, China). The wild-type sequence of STAT3-3′-UTR (STAT3-3′-UTR-wt) and the mutant sequence of STAT3-3′-UTR without miR-874 binding site (STAT3-3′-UTR-mut) were designed. The corresponding luciferase reporter vector was constructed. MiR-874 mimics were used for transfection of cardiomyocytes. Sequences of miR-874 mimics were as follows: antisense strand: 5′-P-UAUGGCUU(5-N-U)UCAUUCCUAUGUGA-Biotin-3′; sense strand: 5′-P-UCACAUAGGAAUGAAAAGCCAUA-3′. Luciferase activity was analyzed using a dual luciferase reporter gene assay kit (Promega, Madison, WI). Forty-eight hours after transfection, the previous medium was discarded and cells were washed with phosphate buffer twice. Each well was added with 100 μl of passive lysis buffer, shaken gently for 15 min at room temperature, and then cell lysate was collected. The prereading time of the program was set as 2 s and the reading value was 10 s, and the sampling amount of LARII, Stop & Glo® Reagent (Promega) was 100 μl. The prepared LARII, Stop & Glo® Reagent were added onto a luminous tube with cell lysate (20 μl in each sample). The solution was detected using a chemiluminescence detector (Modulus™; Turner BioSystems Inc., Sunnyvale, CA) whose longest luminous wavelength was 560 nm. The experiment was repeated three times in each group and its mean value was taken.
2.7 Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
The total RNA in the tissue samples was extracted by TRIzol according to the TRIzol reagent manual (Invitrogen Inc., Carlsbad, CA) and then dissolved in ultrapure water treated with diethyl pyrocarbonate (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd., Shanghai, China). The optical density (OD) at 260 and 280 nm wavelength was measured by a ND-1000 UV/Visible Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The quality of total RNA was determined and the RNA concentration was then adjusted to 100 ng/μl. The total RNA was reverse transcribed using a two-step method according to the kit instruction (Thermo Fisher Scientific, San Jose, CA). The reaction conditions were at 70°C for 10 min, ice bath for 2 min, at 42°C for 60 min, and at 70°C for 10 min. The complementary DNA obtained by reverse transcription was stored temporarily in a −80°C refrigerator. The SYBR Green method was adopted when performing RT-qPCR. The reaction system was prepared according to the reagent kit (Marrone Bio Innovations, Davies, CA) manual: SYBR Green mix 12.5 μl, forward primer 1 μl, reverse primer 1 μl, DNA template 1–4 μl, and brought up to 25 μl with ddH2O. The primer sequences are shown in Table 1. The RT-qPCR condition was one cycle of predenaturation at 95°C for 30 s, 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 20 s, and extension at 70°C for 10 s. The reaction was performed using a RT-qPCR instrument platform (Bio-Rad iQ5; Bio-Rad Laboratories, Hercules, CA). U6 was used as internal reference for miR-874 and β-actin for other genes. The expression of each target gene was calculated using relative quantitative method. Each experiment was repeated three times and the mean value was obtained.
| Genes | Primer sequences (5′–3′) |
|---|---|
| miRNA-874 | F: TGCGGCGGCCCCACGCACCAG |
| R: CCAGTGCAGGGTCCGAGGT | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCAGAATTTGCGT | |
| JAK2 | F: GCAAAGGTAACTTCGGGAGTGT |
| R: AGTCTCGGAGGTGCTCTTCAG | |
| STAT3 | F: GAAACAACCAGTCTGTGACCAG |
| R: CACGTACTCCATTGCTGACAAG | |
| Bax | F: CTGAGCTGACCTTGGAGC |
| R: GACTCCAGCCACAAAGATG | |
| Bcl-2 | F: TGCACCTGAGCGCCTTCAC |
| R: TAGCTGATTCGACCATTT | |
| β-actin | F: GACGGCCAGGTCATCACTATTG |
| R: AGGAAGGCTGGAAAAGAGCC |
- Note. Bax: Bcl-2 associated X protein B; Bcl-2: B-cell lymphoma-2; F: forward; JAK2: Janus kinase 2; R: reverse; RT-qPCR: reverse-transcription quantitative polymerase chain reaction; STAT3: signal transducer and activator of transcription 3; β-actin: cytoskeletal β-actin.
2.8 Western blot analysis
The total protein of animal tissues was extracted and added with 1× sodium dodecyl sulfate lysate and boiled at 100°C for 5 min. Protein samples (20 μl) were obtained and loaded in 12% polyacrylamide gel electrophoresis. After the proteins were transferred to the membrane, the membrane was blocked by Tris-buffered saline Tween-20 (TBST) containing 5% bovine serum albumin (BSA) for 1 hr at room temperature. The blocking solution was discarded and the membrane was placed in a plastic groove. The primary antibodies (anti-mouse p-STAT3, ab32143; anti-mouse p-JAK2, ab195055; anti-mouse STAT3, ab30647; anti-mouse Bax, ab32503; anti-mouse Bcl-2, ab59348; anti-mouse β-actin, ab195055; all purchased from Abcam Inc., Cambridge, MA) were diluted at the rate of 1:1,000 with 5% BSA (with transfer surface upward), placed in a refrigerator at 4°C and shaken and incubated overnight. Next day, the membrane was washed by TBST (3 times × 10 min) and incubated with the diluted secondary antibody (Ab6789; Abcam Inc.) at 4°C. After 4–6 hr of incubation, the membrane was washed with TBST (3 times × 15 min). The chemiluminescence reagent A solution and B solution (Yanhui Bio, Shanghai, China) were mixed at the ratio of 1:1, and uniformly dripped on the nitrocellulose membrane. The membrane was developed. All immunoblot bands were subjected to relative OD analysis. The experiment was repeated three times in each group and the mean value was obtained.
2.9 Statistical analysis
All data were analyzed by statistical software SPSS 21.0 (IBM Corp., Armonk, NY). Measurement data were presented as mean ± standard deviation. The t test was used for comparison between two groups and one-way analysis of variance for comparison among multiple groups. The results were considered as statistically significant when the p < 0.05.
3 RESULTS
3.1 Cardiac function of I/R mice was improved by downregulating miR-874 and activating JAK2/STAT3 signaling pathway
Initially, to explore the effect of miR-874 and JAK2/STAT3 signaling pathway on cardiac function of I/R mice, transthoracic echocardiography was applied to measure LVEF and LVFS. There were no marked differences in LVEF, LVFS, HR, and MAP among each group 30 min before and after 30 min of ischemia (p > 0.05). Compared with the sham group, the control group showed no significant difference in the indexes, while other five groups had significantly diminished LVEF and, LVFS, HR, and MAP at 120 min after reperfusion (all p < 0.05). With the passage of time, the reduction became more and more significant and dropped to the lowest point in T4. There was no significant change in the relevant indexes between T4 and T5. Compared with the I/R group, the SEV, SEV + miR-874 inhibitor, and SEV + miR-874 inhibitor + AG490 groups demonstrated obviously increased LVEF, LVFS, HR, and MAP at 120 min after reperfusion (all p < 0.05), while there were no marked differences in the SEV + AG490 group (p > 0.05). Compared with the SEV group, the SEV + AG490 group demonstrated clearly decreased LVEF, LVFS, HR, and MAP at 120 min after reperfusion (both p < 0.05), while the SEV + miR-874 inhibitor group displayed significantly enhanced LVEF, LVFS, HR, and MAP (p < 0.05), and there was no marked difference in the SEV + miR-874 inhibitor + AG490 group (p > 0.05). Compared with the SEV + miR-874 inhibitor group, the SEV + miR-874 inhibitor + AG490 group illustrated notably decreased LVEF, LVFS, HR, and MAP at 120 min after reperfusion (both p < 0.05; Tables 2-5). All experimental mice had no ventricular aneurysm. The aforementioned results reveal that cardiac function of I/R mice can be improved by downregulated miR-874 and activated JAK2/STAT3 signaling pathway.
| LVEF (%) (mean ± standard deviation) | ||||||
|---|---|---|---|---|---|---|
| Groups | T0 | T1 | T2 | T3 | T4 | T5 |
| Control | 80.34 ± 0.95 | 79.81 ± 1.27 | 78.96 ± 0.83 | 80.13 ± 1.30 | 78.95 ± 1.13 | 78.69 ± 1.37 |
| Sham | 81.13 ± 0.74 | 79.50 ± 0.82 | 80.78 ± 1.30 | 78.97 ± 0.82 | 79.35 ± 1.09 | 80.23 ± 1.52 |
| I/R | 80.63 ± 0.86 | 80.43 ± 0.63 | 55.25 ± 1.61a | 53.33 ± 0.91a | 51.18 ± 0.72a | 55.86 ± 0.84a |
| SEV | 81.02 ± 0.69 | 78.99 ± 1.67 | 70.54 ± 1.23a,b | 67.42 ± 0.76 | 61.38 ± 0.93 | 62.06 ± 1.01a,b |
| SEV + miR-874 inhibitor | 80.96 ± 1.03 | 79.97 ± 1.56 | 54.76 ± 1.03a,c | 52.28 ± 0.75 | 50.96 ± 0.87 | 56.84 ± 0.80a,c |
| SEV + AG490 | 81.11 ± 1.12 | 80.60 ± 0.79 | 75.21 ± 1.10a,b,c | 71.63 ± 1.42 | 67.48 ± 0.81 | 67.90 ± 1.07a,b,c |
| SEV + miR-874 inhibitor + AG490 | 80.87 ± 1.23 | 79.62 ± 1.01 | 71.78 ± 1.24a,b,d | 65.99 ± 0.83 | 62.19 ± 0.57 | 63.11 ± 0.74a,b,d |
- Note. I/R: ischemia–reperfusion; LVEF: left ventricular ejection fraction; SEV: sevoflurane; T0: 30 min before ischemia; T1: 30 min after ischemia; T2: 120 min after reperfusion; T3: 4 hr after reperfusion; T4: 8 hr after reperfusion; T5: 12 hr after reperfusion.
- a p < 0.05 versus the sham group.
- b p < 0.05 versus the I/R group.
- c p < 0.05 versus the SEV group.
- d p < 0.05 versus the SEV + miR-874 inhibitor group.
| LVFS (%) (mean ± standard deviation) | ||||||
|---|---|---|---|---|---|---|
| Groups | T0 | T1 | T2 | T3 | T4 | T5 |
| Control | 42.05 ± 0.98 | 41.35 ± 0.81 | 41.99 ± 1.07 | 42.84 ± 1.27 | 41.36 ± 1.19 | 40.87 ± 1.34 |
| Sham | 42.87 ± 1.09 | 42.30 ± 1.67 | 41.00 ± 0.98 | 41.59 ± 1.30 | 42.05 ± 1.57 | 42.33 ± 1.88 |
| I/R | 42.13 ± 0.74 | 40.76 ± 1.12 | 24.75 ± 0.89a | 22.94 ± 0.83a | 20.86 ± 1.09a | 21.99 ± 0.84a |
| SEV | 43.23 ± 0.60 | 40.98 ± 0.87 | 33.36 ± 1.27a,b | 30.66 ± 0.72a,b | 27.39 ± 0.93a,b | 29.11 ± 1.06a,b |
| SEV + miR-874 inhibitor | 42.67 ± 1.02 | 41.36 ± 1.21 | 25.29 ± 0.75a,c | 23.76 ± 0.90a,c | 20.25 ± 0.74a,c | 23.52 ± 1.09a,c |
| SEV + AG490 | 43.52 ± 0.88 | 41.54 ± 1.56 | 37.67 ± 1.28a,b,c | 35.08 ± 0.91a,b,c | 31.05 ± 1.25a,b,c | 34.28 ± 1.43a,b,c |
| SEV + miR-874 inhibitor + AG490 | 41.81 ± 1.22 | 40.97 ± 1.68 | 32.45 ± 1.16a,b,d | 29.78 ± 1.14a,b,d | 25.62 ± 0.65a,b,d | 27.82 ± 0.90a,b,d |
- Note. I/R: ischemia–reperfusion; LVFS: left ventricular shortening fraction; SEV: sevoflurane; T0: 30 min before ischemia; T1: 30 min after ischemia; T2: 120 min after reperfusion; T3: 4 hr after reperfusion; T4: 8 hr after reperfusion; T5: 12 hr after reperfusion.
- a p < 0.05 versus the sham group.
- b p < 0.05 versus the I/R group.
- c p < 0.05 versus the SEV group.
- d p < 0.05 versus the SEV + miR-874 inhibitor group.
| HR (%) (mean ± standard deviation) | ||||||
|---|---|---|---|---|---|---|
| Groups | T0 | T1 | T2 | T3 | T4 | T5 |
| Control | 439.33 ± 31.21 | 436.50 ± 25.87 | 432.83 ± 28.18 | 434.83 ± 34.10 | 434.17 ± 28.24 | 433.50 ± 25.61 |
| Sham | 435.67 ± 32.93 | 438.00 ± 30.29 | 437.17 ± 29.34 | 433.00 ± 29.59 | 435.83 ± 32.04 | 438.33 ± 26.98 |
| I/R | 429.50 ± 27.41 | 421.17 ± 25.28 | 236.67 ± 12.50a | 211.17 ± 19.11a | 206.00 ± 17.08a | 219.00 ± 16.71a |
| SEV | 433.83 ± 38.64 | 430.67 ± 34.03 | 299.00 ± 22.25a,b | 285.83 ± 13.99a,b | 275.67 ± 21.50a,b | 298.33 ± 17.37a,b |
| SEV + miR-874 inhibitor | 426.00 ± 39.77 | 425.33 ± 31.37 | 227.50 ± 15.07a,c | 213.67 ± 24.13a,c | 199.00 ± 13.04a,c | 224.67 ± 19.62a,c |
| SEV + AG490 | 436.17 ± 35.01 | 432.50 ± 29.60 | 367.17 ± 18.36a,b,c | 358.00 ± 25.08a,b,c | 342.17 ± 17.34a,b,c | 370.00 ± 23.81a,b,c |
| SEV + miR-874 inhibitor + AG490 | 425.00 ± 37.48 | 421.83 ± 28.17 | 306.00 ± 22.80a,b,d | 289.50 ± 20.39a,b,d | 271.33 ± 18.57a,b,d | 293.11 ± 21.07a,b,d |
- Note. HR: heart rate; I/R: ischemia–reperfusion; SEV: sevoflurane; T0: 30 min before ischemia; T1: 30 min after ischemia; T2: 120 min after reperfusion; T3: 4 hr after reperfusion; T4: h8 hr after reperfusion; T5: 12 hr after reperfusion.
- a p < 0.05 versus the sham group.
- b p < 0.05 versus the I/R group.
- c p < 0.05 versus the SEV group.
- d p < 0.05 versus the SEV + miR-874 inhibitor group.
| MAP (mm Hg) (mean ± standard deviation) | ||||||
|---|---|---|---|---|---|---|
| Groups | T0 | T1 | T2 | T3 | T4 | T5 |
| Control | 112.73 ± 8.52 | 105.78 ± 9.50 | 109.94 ± 7.72 | 112.38 ± 8.59 | 111.64 ± 7.52 | 107.85 ± 9.11 |
| Sham | 107.64 ± 9.08 | 109.45 ± 9.33 | 108.99 ± 7.95 | 113.73 ± 8.00 | 113.35 ± 9.40 | 111.46 ± 10.38 |
| I/R | 112.30 ± 7.95 | 98.37 ± 7.36 | 53.56 ± 3.08a | 51.86 ± 4.17a | 44.71 ± 5.27a | 46.37 ± 4.52a |
| SEV | 106.94 ± 8.13 | 103.51 ± 6.03 | 71.30 ± 5.83a,b | 69.74 ± 4.80a,b | 65.38 ± 3.92a,b | 68.53 ± 5.81a,b |
| SEV + miR-874 inhibitor | 99.59 ± 9.10 | 97.51 ± 6.24 | 52.09 ± 4.16a,c | 50.90 ± 4.08a,c | 44.85 ± 3.86a,c | 47.22 ± 3.93a,c |
| SEV + AG490 | 107.84 ± 10.33 | 98.42 ± 8.11 | 90.10 ± 5.92a,b,c | 87.81 ± 6.17a,b,c | 84.05 ± 7.63a,b,c | 88.05 ± 4.16a,b,c |
| SEV + miR-874 inhibitor + AG490 | 110.62 ± 9.18 | 95.81 ± 9.55 | 72.69 ± 6.51a,b,d | 70.35 ± 4.69a,b,d | 64.41 ± 5.38a,b,d | 67.11 ± 5.09a,b,d |
- Note. I/R: ischemia–reperfusion; MAP: mean arterial pressure; SEV: sevoflurane; T0: 30 min before ischemia; T1: 30 min after ischemia; T2: 120 min after reperfusion; T3: 4 hr after reperfusion; T4: 8 hr after reperfusion; T5: 12 hr after reperfusion.
- a p < 0.05 versus the sham group.
- b p < 0.05 versus the I/R group.
- c p < 0.05 versus the SEV group.
- d p < 0.05 versus the SEV + miR-874 inhibitor group.
3.2 Downregulated miR-874 and activated JAK2/STAT3 signaling pathway ameliorate pathological changes of myocardium in I/R mice
Then, H&E staining was carried out to explore the pathological changes of myocardium in I/R mice. In the control and sham groups, the arrangement of the myocardial fibers was neat and tight and arranged in bundles. The morphology of cardiomyocytes was normal without interstitial edema in cells. The nuclear membrane was complete with uniform distribution of chromatin. Compared with the sham group, in the I/R group, the arrangement of myocardial fibers was disordered; the cells were swelled and the interstitial edema in cells was obvious; the boundaries between cells were blurred, most of the stripes disappeared and the cardiomyocytes were largely necrotic with significantly elevated myocardium damage score. Compared with the I/R group, in the SEV group, the arrangement of myocardial fibers was relatively neat, and the necrosis range of cardiomyocytes and the myocardium damage score were significantly reduced. Compared with the SEV group, in the SEV + AG490 group, the arrangement of myocardial fibers was disordered and the necrosis range of cardiomyocytes and the myocardium damage score were increased; however, in the SEV + miR-874 inhibitor group, the arrangement of myocardial fibers was relatively neat, the cells were in mild edema, and the necrosis range of myocardial cells and the myocardium damage score were notably reduced. Compared with the SEV + AG490 group, in the SEV +miR-874 inhibitor + AG490 group, the arrangement of myocardial fibers was disordered and the necrosis range of cardiomyocytes and the myocardium damage score were significantly increased (Figure 1a,b). The above results demonstrate that miR-874 inhibition and JAK2/STAT3 signaling pathway activation is responsible for improved pathological changes of myocardium in I/R mice
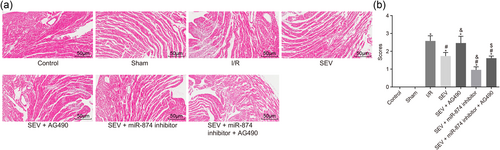
H&E staining reveals that miR-874 inhibition and JAK2/STAT3 signaling pathway activation improve pathological changes of myocardium in I/R mice (×200). (a) H&E staining of pathological changes of myocardium. (b) Quantitative analysis indicates myocardium damage score was downregulated with treatment of miR-874 inhibitor and JAK2/STAT3 signaling pathway activator. *p < 0.05 versus the sham group; #p < 0.05 versus the I/R group; &p < 0.05 versus the SEV group; $p < 0.05 versus the SEV + AG490 group; data were expressed as mean ± standard deviation; the one-way ANOVA was performed for statistical analysis; and the experiment was repeated three times. ANOVA: analysis of variance; H&E: hematoxylin–eosin; I/R: ischemia–reperfusion; JAK2: Janus kinase 2; miR-874: microRNA-874; SEV: sevoflurane; STAT3: signal transducers and activators of transcription 3 [Color figure can be viewed at wileyonlinelibrary.com]
3.3 Downregulated miR-874 and activated JAK2/STAT3 signaling pathway repress myocardial apoptosis
Subsequently, TUNEL staining was applied to investigate the effect of miR-874 and JAK2/STAT3 signaling pathway on myocardial apoptosis. Cells with brown granules in the nucleus were considered as apoptotic cells (Figure 2a,b). There was no obvious apoptosis in the control and sham groups. Compared with the sham group, the I/R group showed a large number of apoptotic cardiomyocytes (p < 0.05). Compared with the I/R group, the SEV, SEV + miR-874 inhibitor, and SEV + miR-874 inhibitor + AG490 groups demonstrated decreased significantly AI (all p < 0.05), while there was no marked difference in the SEV + AG490 group (p > 0.05). Compared with the SEV group, the SEV + AG490 group had significantly higher AI (p < 0.05), and the SEV + miR-874 inhibitor group showed notably lower AI (p < 0.05), while there was no marked difference between the SEV + miR-874 inhibitor + AG490 group (p > 0.05). Compared with the SEV + miR-874 inhibitor group, the SEV + miR-874 inhibitor + AG490 group illustrated evidently higher AI (p < 0.05), indicating decreased miR-874 and activated JAK2/STAT3 signaling pathway inhibits myocardial apoptosis

TUNEL staining demonstrates that miR-874 inhibition and JAK2/STAT3 signaling pathway activation decreases myocardial apoptosis in I/R mice (×400). (a) TUNEL staining reflecting apoptotic cardiomyocytes. (b) AI of cardiomyocytes. *p < 0.05 versus the sham group; #p < 0.05 versus the I/R group; &p < 0.05 versus the SEV group; $p < 0.05 versus the SEV + AG490 group; data were expressed as mean ± standard deviation; the one-way ANOVA was performed for statistical analysis; and the experiment was repeated three times. ANOVA: analysis of variance; AI: apoptosis index; I/R: ischemia–reperfusion; JAK2: Janus kinase 2; miR-874: microRNA-874; SEV: sevoflurane; STAT3: signal transducers and activators of transcription 3; TUNEL, terminal deoxynucleotidyl transferase (TdT)-mediated dUTP biotin nick end labeling [Color figure can be viewed at wileyonlinelibrary.com]
3.4 STAT3 is a target gene of miR-874
There was a specific binding region between the STAT3-3′-UTR 864–871 sequence and the miR-874 sequence by prediction from the online analytical software, indicating that STAT3 was a target gene of miR-874 (Figure 3a). The dual luciferase reporter gene assay was applied to identify whether STAT3 was a target of miR-874 (Figure 3b). MiR-874 mimics had no marked effect on the luciferase activity intensity of STAT3-3′-UTR-mut (p > 0.05), but the activity of luciferase in STAT3-3′-UTR-wt significantly decreased (p < 0.05), suggesting that miR-874 could specifically bind STAT3-3′-UTR and downregulate the expression of STAT3 gene after transcription.
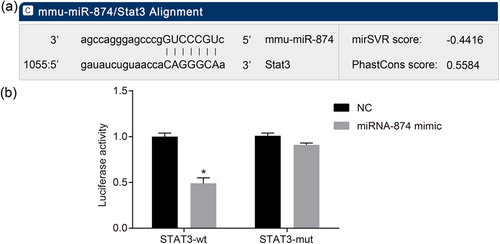
MiR-874 negatively regulates STAT3. (a) Binding regions between STAT3-3′-UTR and miR-874 sequence. (b) Luciferase activity of the STAT3 wt and TRAIL mut after transfection. *p < 0.05 versus the sham group; #p < 0.05 versus the I/R group; &p < 0.05 versus the SEV group; $p < 0.05 versus the SEV + AG490 group; data were expressed as mean ± standard deviation; the one-way ANOVA was performed for statistical analysis; and the experiment was repeated three times. ANOVA: analysis of variance; I/R: ischemia–reperfusion; JAK2: Janus kinase 2; miR-874: microRNA-874; mut: mutant type; SEV: sevoflurane; STAT3: signal transducers and activators of transcription 3; wt: wild type; 3′-UTR: 3′-untranslated region [Color figure can be viewed at wileyonlinelibrary.com]
3.5 MiR-874 is upregulated in myocardial tissues of I/R mice
RT-qPCR was used to determine miR-874 expression, thereby investigating the effect of miR-874 on I/R. There was no significant difference in miR-874 expression between the control and sham groups. Compared with the sham group, other five groups had significantly increased miR-874 expression (all p < 0.05). Compared with the I/R group, the SEV, SEV + AG490, SEV + miR-874 inhibitor, and SEV + miR-874 inhibitor + AG490 groups demonstrated significantly decreased miR-874 expression (all p < 0.05). Compared with the SEV group, the SEV + miR-874 inhibitor group displayed significantly decreased miR-874 expression (p < 0.05). There was no marked difference in miR-874 expression between the SEV + miR-874 inhibitor group and the SEV + miR-874 inhibitor + AG490 group (p < 0.05; Figure 4). These results display that downregulated miR-874 correlates to I/R.
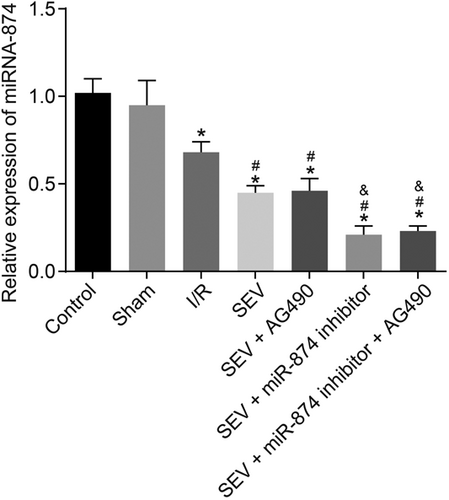
MiR-874 is upregulated in mice with I/R. *p < 0.05 versus the sham group; #p < 0.05 versus the I/R group; &p < 0.05 versus the SEV group; data were expressed as mean ± standard deviation; the one-way ANOVA was performed for statistical analysis; and the experiment was repeated three times. ANOVA: analysis of variance; I/R: ischemia–reperfusion; JAK2: Janus kinase 2; miR-874: microRNA-874; SEV: sevoflurane; STAT3: signal transducers and activators of transcription 3
3.6 MiR-874 represses the activation of JAK2/STAT3 signaling pathway
Then, to explore the effect of miR-874 on the JAK2/STAT3 signaling pathway, RT-qPCR and western blot analysis were conducted to measure the expression of the JAK2/STAT3 signaling pathway-related genes. There was no significant difference in the JAK2/STAT3 signaling pathway-related genes between the control and sham groups. Compared with the sham group, the other five groups displayed increased significantly mRNA and protein levels of JAK2 and STAT3 (all p < 0.05). Compared with the I/R group, the SEV, SEV + miR-874 inhibitor and SEV + miR-874 inhibitor + AG490 groups illustrated evidently increased mRNA expression of JAK2 and STAT3 as well as levels of p-JAK2/t-JAK2 and p-STAT3/t-STAT3 (p < 0.05), but there were no marked differences in the SEV + AG490 group (p > 0.05). Compared with the SEV group, the SEV + AG490 group demonstrated notably decreased mRNA expression of JAK2 and STAT3 as well as levels of p-JAK2/t-JAK2 and p-STAT3/t-STAT3(p < 0.05), but the SEV + miR-874 inhibitor group illustrated increased significantly mRNA expression of JAK2 and STAT3 as well as levels of p-JAK2/t-JAK2 and p-STAT3/t-STAT3(p < 0.05), and no marked differences were observed in the SEV + miR-874 inhibitor + AG490 group (p > 0.05). Compared with the SEV + miR-874 inhibitor group, the SEV + miR-874 inhibitor + AG490 group showed evidently decreased mRNA expression of JAK2 and STAT3 as well as levels of p-JAK2/t-JAK2 and p-STAT3/t-STAT3 (p < 0.05; Figure 5a–c). The aforementioned results demonstrate that the JAK2/STAT3 signaling pathway is inhibited by miR-874.
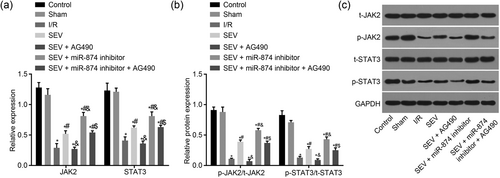
MiR-874 inhibits the JAK2/STAT3 signaling pathway. (a) mRNA levels of JAK2 and STAT3 in response to the treatment of SEV, SEV + miR-874 inhibitor, SEV + AG490, SEV + miR-874 inhibitor + AG490. (b) The extent of JAK2 and STAT3 phosphorylation in response to the treatment of SEV, SEV + miR-874 inhibitor, SEV + AG490, SEV + miR-874 inhibitor + AG490. (c) The gray value of p-JAK2 and p-STAT3 protein bands in response to the treatment of SEV, SEV + miR-874 inhibitor, SEV + AG490, SEV + miR-874 inhibitor + AG490. *p < 0.05 versus the sham group; #p < 0.05 versus the I/R group; &p < 0.05 versus the SEV group; $p < 0.05 versus the SEV + AG490 group; data were expressed as mean ± standard deviation; the one-way ANOVA was performed for statistical analysis; and the experiment was repeated three times. ANOVA: analysis of variance; I/R: ischemia–reperfusion; JAK2: Janus kinase 2; miR-874: microRNA-874; mRNA: messenger RNA; SEV: sevoflurane; STAT3: signal transducers and activators of transcription 3
3.7 Downregulated miR-874 suppresses myocardial apoptosis by regulating the apoptosis-related genes
Finally, Bcl-2 associated X protein B (Bax) and B-cell lymphoma-2 (Bcl-2) expression was determined by RT-qPCR and western blot analysis to further explore the effect of miR-874 on myocardial apoptosis. There was no significant difference in the apoptosis-related genes between the control and sham groups. Compared with the sham group, the other five groups had significantly increased expression of Bax and sharply decreased expression of Bcl-2 (p < 0.05). Compared with the I/R group, in the SEV, SEV + miR-874 inhibitor and SEV + miR-874 inhibitor + AG490 groups, Bax mRNA and protein expression decreased observably (p < 0.05) and Bcl-2 mRNA and protein expression were enhanced evidently (p < 0.05), while in the SEV + AG490 group, there was no marked difference in the Bcl-2 mRNA and protein expression (p > 0.05). Compared with the SEV group, in the SEV + AG490 group, the Bax mRNA and protein expression obviously increased, while Bcl-2 mRNA and protein expression sharply decreased (p < 0.05). In the SEV + miR-874 inhibitor group, the Bax mRNA and protein expression markedly decreased and the Bcl-2 mRNA and protein expression remarkably increased (p < 0.05). There was no marked change in the SEV + miR-874 inhibitor + AG490 group (p > 0.05). Compared with the SEV + miR-874 inhibitor group, in the SEV + miR-874 inhibitor + AG490 group, the Bax mRNA and protein expression obviously increased and the mRNA and protein expression of Bcl-2 sharply decreased (p < 0.05; Figure 6a–c), indicating that miR-874 inhibition is responsible for decreased myocardial apoptosis.
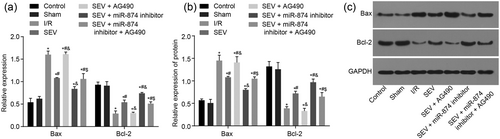
MiR-874 inhibition suppresses myocardial apoptosis by regulating the apoptosis-related genes. (a) mRNA levels of Bax and Bcl-2 in response to the treatment of SEV, SEV + miR-874 inhibitor, SEV + AG490, SEV + miR-874 inhibitor + AG490. (b) The protein levels of Bax and Bcl-2 in response to the treatment of SEV, SEV + miR-874 inhibitor, SEV + AG490, SEV + miR-874 inhibitor + AG490. (c) The gray value of Bax and Bcl-2 protein bands in response to the treatment of SEV, SEV + miR-874 inhibitor, SEV + AG490, SEV + miR-874 inhibitor + AG490. *p < 0.05 versus the sham group; #p < 0.05 versus the I/R group; &p < 0.05 versus the SEV group; $p < 0.05 versus the SEV + AG490 group; data were expressed as mean ± standard deviation; the one-way ANOVA was performed for statistical analysis; and the experiment was repeated three times. ANOVA: analysis of variance; Bax: Bcl-2-associated X protein; Bcl-2: B-cell lymphoma-2; I/R: ischemia–reperfusion; JAK2: Janus kinase 2; miR-874: microRNA-874; mRNA: messenger RNA; SEV: sevoflurane; STAT3: signal transducers and activators of transcription 3
4 DISCUSSION
The postgenome sequencing era witnessed a significant theoretical breakthrough of the discovery of noncoding RNA in the human genome, and thus improved understanding of its essential for the continued development of cancer research (Nohata et al., 2013). As a leading cause of death, the deleterious effects of myocardial I/R have been studied over the past four decades (Benhabbouche, Crola da Silva, Abrial, & Ferrera, 2011). This study was supposed to explore the effect of miR-874 on myocardial I/R injury in mice models pretreated with SEV by targeting STAT3 through the JAK2/STAT3 signaling pathway. Consequently, we found that miR-874 downregulation inhibits cardiomyocytes apoptosis in mice with myocardial I/R injury via the JAK2/STAT3 pathway by downregulating STAT3
The study indicated that miR-874 expression was higher in I/R mice and decreased in I/R mice following SEV pretreatment, and downregulated miR-874 activates the JAK2/STAT3 signaling pathway, thus improving myocardial I/R injury in mice. MiR-126, miR-1, and miR-133a have been proved to protect against myocardial cell apoptosis caused by I/R injury (He et al., 2011; B. Li, Tao, & Huang, 2015). Moreover, suppression of miR-135b-5p prevents myocardial I/R injury through the JAK2/STAT3 signaling pathway by mediating the inhalation anesthesia with SEV has been previously reported (X. J. Xie et al., 2017). JAK2/STAT3 signaling pathway has been proved to specifically associate with preventing myocardial I/R (Y. Yang et al., 2013). The activation of the JAK/STAT signaling pathway could upregulate inducible nitric oxide synthase through ischemic preconditioning, thus helping adaption of the heart to ischemic stress (Das, Salloum, Durrant, Ockaili, & Kukreja, 2012). In addition, the luciferase reporter gene assay showed that miR-874 could specifically bind to STAT3-3′-UTR and regulate STAT3 gene. MiRNAs bind to 3′-UTRs of target genes and regulate gene expression at posttranscriptional level by reducing the stability of nascent mRNA strands, protein translation, or both, and resulting in the reduction of target protein productions (Wu et al., 2013). As a transcription factor, STAT3 regulates a series of genes involved in the regulation of various biological processes and the STAT3 signaling pathway is thought to be closely related to miRNAs (Cao et al., 2013).
We evidenced that downregulated miR-874 inhibits cardiomyocyte apoptosis in mice with myocardial I/R injury and reduces the expression of Bax and increases expression of Bcl-2. Apoptosis may be caused in susceptible cells through various normal physiological stimuli, together with the harmful environment and cytotoxic agents and common inducers of apoptosis containing Ca2+ and oxygen free radicals or oxidative stress, which are also involved in the pathogenesis of myocardial I/R injury (Galang, Sasaki, & Maulik, 2000). Cardiomyocyte apoptosis together with inflammatory reaction are believed to be hallmarks of myocardial I/R injury; moreover, it is reported that cardiomyocyte apoptosis emerges shortly after ischemia, and is magnified after reperfusion, leading to cardiomyocyte death (Guo et al., 2013). As cytosolic proteins, Bcl-2 targets the nucleus and inhibits apoptosis with a lipid-anchoring domain, while Bax, one of the members of the Bcl family, homodimerizes and takes shape heterodimers with Bcl-2 protein and reduces antiapoptotic effects of Bcl-2 leading to apoptotic death (Z. Xie et al., 2001). Reduced apoptosis, decreased Bax expression, and increased expression of Bcl-2/Bax indicate attenuated I/R injury (Ji, Yue, Wu, & He, 2004). Inhibition of STAT3 has been shown to increase apoptosis of cardiomyocytes and infarct size after I/R and cause loss of protection during ischemic postconditioning and pharmacological preconditioning (Das et al., 2012). Moreover, in a current study, the protective effect of sRAGE on cardiomyocytes from apoptosis caused by I/R was related with the activation of the JAK2/STAT3 signaling pathway, where increased protein expression of the extent of STAT3 and JAK2 phosphorylation were observed (X. Jiang et al., 2015). It has been demonstrated that upregulation of miR-24 reduces expression of BIM, decreases cardiomyocyte apoptosis, and thus improving cardiac function after acute myocardial infarction (Pan, Wang, Ling, & Gong, 2017). In addition, miR-106b and miR-15b may function as robust regulators in apoptosis or angiogenesis following myocardial infarction, thus playing essential roles in pathological processes of myocardial infarction (Liu et al., 2012). And miR-7a/b is also proved to protect against hypoxia-induced injury as well as cardiac remodeling in H9c2 cardiomyoblasts involved in Sp1 and PARP-1 (R. Li et al., 2016). In addition, miR-133b and miR-135a induce apoptosis in vitro via the JAK2/STAT3 signaling pathway and functionally counteract oncogenic signaling pathways in clear cell renal cell carcinoma (Zhou, Bi, Gao, & Sun, 2016). MiR-375 mediates the JAK2/STAT3 signaling pathway affecting the expression of Bcl-2 and TWIST1, and thus providing a potential therapeutic target for inflammation-related cancers (Miao, Liu, Xie, Xing, & Xi, 2014). The specific mechanism of miR-874 in alleviating I/R injury is demanded to be clarified in further studies in the future.
Taken together, our study provided evidence that miR-874 inhibition could alleviate myocardial I/R injury by targeting STAT3 through the JAK2/STAT3 signaling pathway. Nevertheless, further studies are required to figure out the specific mechanisms of miR-874 in ischemic heart diseases.
ACKNOWLEDGMENTS
This study is supported by the Youth Medical Talent of Jiangsu Province (QNRC2016464) and 2017 Medical Science and Technology Development Program of Yancheng City (YK2017120, YK2017121, YK2017122). The authors want to show their appreciation to reviewers for their helpful comments.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.