Basal progenitor cells bridge the development, malignant cancers, and multiple diseases of esophagus
Abstract
The esophagus is a pivotal organ originating from anterior foregut that links the mouth and stomach. Moreover, its development involves precise regulation of multiple signal molecules and signal transduction pathways. After abnormal regulation of these molecules in the basal cells of the esophagus occurs, multiple diseases, including esophageal atresia with or without tracheoesophageal fistula, Barrett esophagus, gastroesophageal reflux, and eosinophilic esophagitis, will take place as a result. Furthermore, expression changes of signal molecules or signal pathways in basal cells and the microenvironment around basal cells both can initiate the switch of malignant transformation. In this review, we highlight the molecular events underlying the transition of normal development to multiple esophageal diseases. Additionally, the animal models of esophageal development and related diseases, challenges, and strategies are extensively discussed.
Abbreviations
-
- BE
-
- Barrett's esophagus
-
- BMP
-
- bone morphogenetic protein
-
- EA
-
- esophageal atresia
-
- EAC
-
- esophageal adenocarcinoma
-
- EE
-
- esophageal epithelium
-
- EoE
-
- eosinophilic esophagitis
-
- ESCC
-
- esophageal squamous cell carcinoma
-
- GEJ
-
- gastro-esophageal junction
-
- GERD
-
- gastro-esophageal reflux disease
-
- LES
-
- lower esophageal sphincter
-
- SCJ
-
- squamocolumnar junction
-
- Shh
-
- sonic hedgehog
-
- TEF
-
- tracheoesophageal fistula
1 BACKGROUND
The esophagus is a pivotal organ that links the mouth to the stomach; it is important to transport nutrition and food in the embryo. Similar to the trachea, the esophagus also derives from anterior foregut. Along with coordinated interplay of the epithelium and mesenchyme, separation of the single anterior foregut tube into two different tubes also requires normal regulation of multiple signal pathways and transcription factors. Transcription factors and signaling molecules play their key roles in establishing the dorsal-ventral expression pattern, and disruption of their expression patterns leads to esophageal atresia with or without tracheoesophageal fistula (EA/TEF). Previous studies have reviewed the genetic and cellular mechanisms that are involved in the development of anterior foregut and esophagus (Jacobs, Ku, & Que, 2012; Que, Choi, Ziel, Klingensmith, & Hogan, 2006).
After applications of animal models in analyzing esophageal diseases, abnormalities in basal cells residing in the stratified squamous epithelium have been demonstrated to be closely correlated with multiple esophageal diseases, including Barrett esophagus, gastroesophageal reflux disease (GERD), eosinophilic esophagitis (EoE), esophageal squamous cell carcinoma (ESCC), and esophageal adenocarcinoma (EAC). In this review, we will discuss these esophageal diseases and highlight the abnormalities in basal cells responsible for esophageal cancer.
2 OVERVIEW OF ESOPHAGUS DEVELOPMENTAL PROCESS AND THE BASAL CELLS IN THE ESOPHAGUS
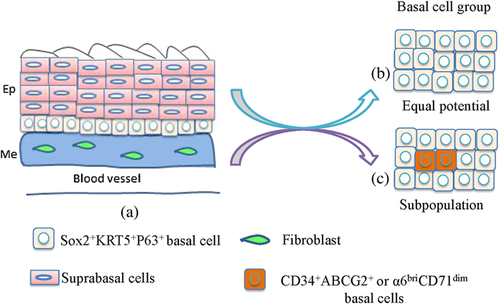
The developmental process of the anterior foregut in the mammalian embryo involves changes in the behavior of both the epithelial endoderm and mesoderm. Morphogenetic processes that occur include the extrusion of midline notochord cells from the epithelial definitive endoderm, the folding of the endoderm into a foregut tube, and the subsequent separation of the foregut tube into the trachea and esophagus (Que et al., 2006). During murine early development, the ventral fold of the endodermal epithelial sheet during gastrulation at approximately embryonic day 8 (E8.0) forms the foregut tube. After formation of the foregut tube, dorsal-ventral patterning of signaling molecules and transcription factors in both the epithelium and the surrounding mesenchyme are also established, and a true expression pattern is required for normal anterior foregut morphogenesis (E9.5-E11.5). All of these events result in generation of the esophagus from the dorsal region of the foregut and production of tracheal and lung buds from the ventral region (Jacobs et al., 2012). At the time of foregut separation (∼ E11.0), the lumen is comprised of a ciliated simple columnar epithelium. It is then gradually replaced by a stratified squamous epithelium that consists of an undifferentiated basal progenitor layer and several differentiated suprabasal layers (Fig. 3B) (Jacobs et al., 2012; Yu et al., 2005). Meanwhile, the mesenchymal cells surrounding the nascent esophagus proliferate and differentiate into multiple layers of muscle cells (Jacobs et al., 2012). Therefore, after the separation of the esophagus from the foregut, the esophagus is ensheathed by layers of muscle and lined with stratified squamous epithelium (Figure 1a).
During the normal development of mouse esophagus, a key step is the conversion of single columnar epithelium to stratified squamous epithelium. By using esophageal explant model isolated from E11.5 mouse embryos, occurrence of a conversion of columnar to stratified squamous epithelium is confirmed based on the expression of specific genes. At E11.5, mouse embryonic esophagus is lined with a simple single columnar epithelium surrounded by undifferentiated mesenchyme after the foregut separates into the esophagus and trachea, and some of the cells located in the basal layer of stratified squamous epithelium directly originate from the former columnar cells and DNA methylation is required for the loss of K8 expression (Yu et al., 2005).
In the adult, basal cells in the stratified squamous epithelium of esophagus are stem cells, and they can self-renew and differentiate into suprabasal layers. However, controversies about their homogeneity or heterogeneity still occur. No slow-cycling or quiescent epithelial stem cells are found in the esophageal epithelium of mouse, and the esophageal epithelium is maintained by a single progenitor cell population that divides randomly to generate proliferating and differentiating progeny with equal probability, and this population can also reversibly switch between homeostatic and regenerative behavior to respond injury (Doupe et al., 2012) (Figure 1b). Yet, other evidences strongly suggest that only a subpopulation of basal cells is stem cell (Figure 1c). In human esophageal epithelium, the epithelial basal layer consists of two distinct regions, one is papillary basal layer (PBL) overlying the papillae of the supporting connective tissue, and the other is interpapillary basal layer (IBL) covering the interpapillary zone. Moreover, the proliferation in PBL is more frequent then in IBL through Ki-67 staining experiment; few basal cells in IBL are Ki-67 positive and mitotic figures in PBL are four times more common than in IBL. In addition, the orientation of individual mitoses in PBL cells is symmetrical while asymmetrical division occurs in IBL. Hence, all these data suggest that the IBL is rich for stem cells for the causes of their infrequent and asymmetrical divisions, large and active clones in culture, and transit amplifying cells locate in the PBL and epibasal layers, and suprabasal cells can not divide any longer except of differentiation (Seery & Watt, 2000).
Additionally, a side population possessing properties of stem cells in mouse esophageal epithelium has been identified using multiple methods. These stem cells belong to a subpopulation of basal cells, and they can also evolve to undifferentiated and differentiated cells to heal wounds (Kalabis et al., 2008). Using cell kinetic studies and the differential expression of multiple cell surface markers, four different populations expressing different levels of surface markers in primary cell population of mouse esophageal mucosa are distinguished as follows: α6briCD71dim, α6briCD71bri, α6dimCD71bri, and CD71+++, and they are designated as a putative esophageal stem cell population, transit-amplifying population, population destined to leave the basal layer and differentiate, and suprabasal cells in the process of differentiation, respectively (Croagh, Phillips, Redvers, Thomas, & Kaur, 2007), indicating that these basal cells in the esophageal epithelium are organized in a hierarchical fashion rather than a random fashion, and exquisite balance among these molecular signals in basal cells is requisite for esophagus development.
3 REGULATION OF SIGNAL MOLECULES IN THE NORMAL ESOPHAGUS DEVELOPMENT AND DISEASES
Many signal molecules participate in the complicated conversion of epithelial cells, and abnormal regulation of these molecules results in defective separation and malformation of the trachea and esophagus. Among these signaling pathways, bone morphogenetic protein (BMP), Wnt, and nuclear respiratory factor (Nrf)2/keap oxidative stress pathways are predominant pathways that regulate the conversion of single columnar epithelium to multiple layers of squamous epithelium.
BMP signaling and the distribution of Bmp molecules are important for the foregut separation using in situ hybridization in section. In the unseparated foregut tube, Bmp4 is preferentially expressed in the ventral mesenchyme, whereas Bmp7 and Bmp inhibitor Noggin are enriched in the dorsal endoderm (Que et al., 2006). Furthermore, transcripts of BMP signaling components and various antagonists are also localized using a BMP reporter mouse line harboring a BRE-lacZ allele and in situ hybridization method. After application of a Sonic hedgehog (shh)-Cre allele to achieve gain- or loss-of-function of Bmpr1a (Alk3) in the embryonic foregut epithelium, high levels of ectopic BMP signaling stall the transition from simple columnar to multilayered undifferentiated epithelium in the esophagus and forestomach, whereas conditional deletion of BMP receptor allows the formation of a multilayered squamous epithelium that cannot differentiate (Rodriguez et al., 2010).
Wnt signaling pathways are highly conserved and they contain three classic signal pathways, and the canonical Wnt/β-catenin pathway and noncanonical Wnt signaling pathway both exert their roles in foregut morphogenesis and separation. Before and after foregut separation, a dynamic pattern of molecules involved in canonical Wnt/β-catenin pathway is confirmed using BAT-Gal reporter line. At E9.5, high activity of Wnt is found in the epithelium of ventral foregut, and their activities can be sustained after the separation of the anterior foregut into the trachea and esophagus at E11.5. Moreover, expression patterns of Wnt2 and Wnt2b from ventral sides to the mesenchyme of ventral foregut are limited to the epithelium, suggesting the Wnt signal receiving cells are located in the epithelium (Jacobs et al., 2012). Furthermore, deletion of Wnt signal molecules leads to the advent of multiple phenotypes including abnormal separation of the foregut tube and lung agenesis, and reduced proliferation, whereas activation of these molecules can reprogram esophagus and stomach endoderm to a lung progenitor fate (Goss et al., 2009). Barx1, a homeobox gene that is highly expressed in the mesenchyme, which is adjacent to the groove for future separation of trachea and esophagus, can inhibit Wnt signaling activity in mouse thoracic foregut and confine it to the ventral region before the trachea and esophagus separation; moreover, Barx1 abrogation leads to changed expression pattern of Wnt activity and Nkx2.1 and severe tracheoesophageal septation defects (Woo, Miletich, Kim, Sharpe, & Shivdasani, 2011). Many cellular processes including cytoskeletal rearrangement and cell shape changes in foregut morphogenesis are also regulated by the noncanonical Wnt signaling. Rhou, a guanosine triphosphatase that acts as the upstream regulator of the Wnt5a/JNK/PCP pathway, is expressed in the ventral and lateral foregut endoderm and its downregulation disrupts the differentiation and morphogenesis of foregut derivatives in cultured embryos; moreover, downregulation of Rhou in individual endoderm cells not only leads to the reduced accumulation of F-actin in the apical territory of epithelium and influences cellular morphology and cytoskeletal organization, but also destroys the conversion of simple columnar epithelium to multilayered squamous epithelium (Loebel et al., 2011).
Nrf2 is a ubiquitous protein belonging to the small family of basic leucine zipper transcription factors, and it is a major regulator of cytoprotective genes encoding detoxification and antioxidant enzymes. Interestingly, expression and activation of Nrf2 both are confirmed in the epithelium of humans and mice; Nrf2 deficiency and GERD in mice, alone or in combination, lead to reduced transepithelial electrical resistance and increased intercellular space in esophageal epithelium. Meanwhile, expression levels of a series of genes and gene sets regulated by Nrf2 also decrease in the esophageal epithelium of Nrf2 knockout mice, along with a significant enhancement of DNA oxidative damage and downregulation of adenosine triphosphate biogenesis, CoxIV and Claudin4, suggesting that Nrf2 deficiency impairs the barrier function of esophageal epithelium and Nrf2 may play a protective role against GERD (Yu et al., 2005).
4 INVOLVEMENT OF BASAL CELLS IN GERD GENERATION
GERD is an esophageal disease caused by the reflux of acid or other stimulus from the stomach; the main reason for the reflux is the incompetence of the antireflux barriers at the gastroesophageal junction (GEJ). The antireflux barriers contain two layers of sphincter: the lower esophageal sphincter (LES) and the crural diaphragm. The gastroesophageal reflux may occur with the advent of lower LES pressure than the intragastric pressure arising from LES hypotension, increased frequency of transient LES relaxation, or enhancement of the intragastric pressure. The severity of GERD progressively increases when reflux occurs during the postprandial period and at night. Moreover, GERD closely correlates with EoE, and in patients with GERD endoscopically visible erosive esophagitis may develop.
Molecular mechanisms underlying both GERD and EoE diseases involve multiple factors including basal cells and inflammatory cytokines. Current studies showed that the abnormal regulation of signal molecules and transcription factors in basal cells can cause abnormal regulation, proliferation, and differentiation of basal cells such as hyperplasia of basal cells; moreover, inflammatory cytokines can also promote the hyperplasia of basal cells, and all of these events finally lead to the occurrence of GERD and EoE. The esophageal epithelium from patients with GERD exhibits lower electrical resistance and higher fluorescein flux than that of normal control patients. GERD is also associated with the cleavage of E-cadherin, and this cleavage may arise from the activation of a membrane metalloproteinase, A Disintegrin And Metalloproteinase (ADAM-10). Moreover, increased junctional permeability was found in the esophageal epithelium of E-cadherin-deleted mouse and in EoE isolated from GERD, suggesting the pivotal roles of E-cadherin in the pathogenesis of GERD (Jovov et al., 2011).
5 CONTRIBUTION OF BASAL CELLS IN BARRETT ESOPHAGUS GENERATION
Barrett esophagus is highlighted by a metaplastic status that stratified squamous epithelium is replaced by simple columnar epithelium. Metaplasia is a process of cell type replacement derived from abnormal differentiation of stem cells or transdifferentiation between mature cell types; it can also protect the esophagus from toxic agents causing chronic inflammation and thus exert more resistance to acid-peptic damage.
So far, four classic hypotheses have been proposed to explain the origin of Barrett esophagus but the precise mechanism underlying this disease remains elusive, and models for the etiology of Barrett esophagus are summarized and depicted in Figure 2. The first hypothesis supports that columnar cells in the epithelium of the GEJ or stomach can migrate and reside in esophagus to reconstitute the gastro-esophageal reflux-damaged squamous epithelium (Bremner et al., 1970) (Figure 2a); however, this hypothesis cannot explain why these columnar epithelial cells only locate in partial esophageal site that is not contiguous with the stomach. The second hypothesis suggests that Barrett esophagus derives from a type of metaplasia called transdifferentiation (Figure 2b). As previously described, a columnar to squamous cell conversion occurs during the development of embryonic esophagus (Yu et al., 2005), conversely, whether a switch that is responsible for the conversion of squamous to columnar cell during Barrett esophagus generation occurs or not is worth validating. The third hypothesis for Barrett esophagus generation is stem cell theory (Figure 2c). When the superficial layers of the esophageal squamous epithelium are damaged by reflux esophagitis, the abnormal differentiation of stem cells locating in the basal layers of squamous esophageal epithelium and/or in the neck region of the esophageal submucosal gland ducts occurs, and the reprogramming of these stem cells eventually leads to Barrett esophagus (Barbera & Fitzgerald, 2010), whereas the exact site of these stem cells and the origin of these metaplastic epithelium are not fully determined.
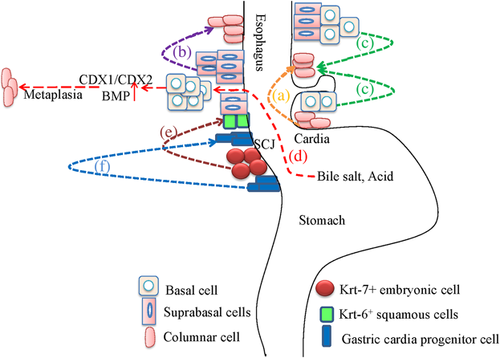
More recently, the fourth hypothesis suggests that Barrett metaplasia involves multiple interactions among developmental signaling pathways, morphogenetic factors, and caudal homeobox (CDX) genes (Figure 2d). In this model, acid, bile, and CDX genes all contribute to the process of Barrett metaplasia. Normally, CDX genes specifically express in the intestine and CDX1/CDX2 uniquely express in the epithelium of the small and large intestine but not that of the esophagus and stomach, and CDX1/CDX2 play their roles not only in the development of gut but also in the generation of intestinal-type morphology and function. Moreover, many studies have demonstrated that the expression of CDX1 or CDX2 can be induced by multiple factors, such as conjugated bile salts, tumor necrosis factor alpha (TNFα) and interleukin(IL)-1β (Wong et al., 2005), cholic and dehydrocholic acids (Kazumori, Ishihara, Rumi, Kadowaki, & Kinoshita, 2006), deoxycholic and chenodeoxycholic acids (Hu et al., 2007), chronic acid exposure (Marchetti, Caliot, & Pringault, 2003), and acid and bile acids (Liu et al., 2007). When expression of CDX genes in the stomach is achieved through genetic manipulation, a phenotype of metaplastic and intestinal epithelium that is similar to that of Barrett esophagus occurs (Souza, Krishnan, & Spechler, 2008). In human embryo, decreased levels of morphogenetic factors which can regulate CDX expression also lead to the conversion of columnar epithelium to squamous epithelium (Que et al., 2006). Furthermore, compared with normal esophagus and stomach, very high levels of CDX1/CDX2 have been confirmed in the biopsy specimens of Barrett metaplasia (Eda et al., 2003; Vallbohmer et al., 2006; Wong et al., 2005), and demethylation of CDX1 is also found in the specialized intestinal metaplasia of Barrett esophagus (Wong et al., 2005).
In rat and human models, activation of the BMP pathway has been proved in esophagitis and Barrett esophagus, and BMP treatment in primary normal squamous esophageal cells also cause the cytokeratin expression pattern of columnar cells, suggesting the BMP pathway may play a role in the conversion of normal esophageal squamous cells to columnar cells (Milano et al., 2007). Conceivably, an increased level of morphogenetic factors may result in the conversion of squamous epithelium to columnar epithelium. Taken together, a pathological model involving multiple factors is proposed. In this model, GERD separates the junctions among squamous cells through acid-peptic damage and then expands the intercellular spaces. These changes lead to the exposure of basal cells to acid, bile salts, and inflammation, which can induce the expression of CDX genes through the demethylation of CDX promoter or increased morphogenetic factors (such as BMP4), and the squamous to columnar cell metaplasia feature of Barrett esophagus forms as a result (Souza et al., 2008) (Figure 2d). Therefore, the chronic inflammation that is principally produced by GERD in basal cells promotes the occurrence of Barrett esophagus.
However, controversies do exist when referring to the involvement of basal cells in Barrett esophagus, and other viewpoints support that the epithelium at the boundary in the stomach contribute to the formation of Barrett esophagus. In p63-deficient mice lacking squamous epithelia, an intestine-like metaplasia possessing similar gene expression profiles to Barrett metaplasia rapidly forms. The Barrett metaplasia arises from a residual embryonic epithelium normally residing at the squamocolumnar junction (SCJ) of adult mice and humans, but does not arise from transdifferentiation, and these Krt-7-positive embryonic cells can migrate to adjacent Krt-6-positive squamous cells after programmed damage to squamous epithelium, which can trigger Barrett metaplasia (Wang et al., 2011) (Figure 2e). In another transgenic mouse model, overexpression of IL-1β in esophagus induces esophagitis, Barrett-like metaplasia and neoplasia, and this process is IL-6 dependent because IL-6 deficiency in L2-IL-1β mice leads to the abolishment of inflammation, metaplasia, and high-grade dysplasia. Moreover, the esophagus of IL-1β mice has similar expression profiles to human Barrett esophagus and EAC, including upregulation of key genes such as BMP4, CDX2, and IL-6. In addition, the progress of Barrett-like metaplasia and dysplasia are promoted by bile acids and/or nitrosamines, and inflammation induced by bile acids and IL-1β leads to the expansion and migration of gastric cardia progenitor cells into the distal esophagus, suggesting Barrett esophagus and EAC may originate from gastric cardia progenitor cells at the SCJ junction (Quante et al., 2012) (Figure 2f).
6 CONTRIBUTION OF BASAL CELLS IN EoE GENERATION
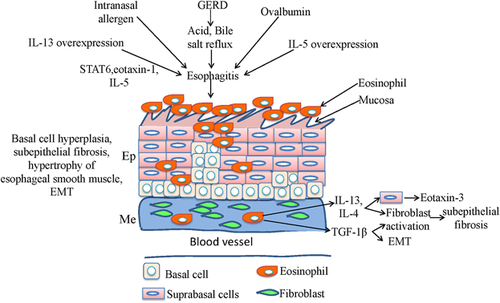
EoE is an immune-mediated syndrome, and the clinical and pathologic traits of EoE comprise dysfunction of the esophagus and hypereosinophilia on esophageal mucosal biopsies. In addition, EoE also involves extensive tissue remodeling that occurs with epithelial hyperplasia, subepithelial fibrosis, and hypertrophy of esophageal smooth muscle. When prolonged exposure of the esophageal mucosa to a pH <4 occurs, mucosal defense will be conquered and severe and complicated esophagitis will ensue as a result; in addition, inflammation in esophageal mucosa may also affect nerves and muscle, which can affect the function of LES and motility of the esophageal body. Moreover, gastric pepsin and duodenal contents caused by GERD will also aggravate the effects of the acid, thereby leading to a more deleterious effect on esophagitis generation.
To determine whether delivery of IL-13 is sufficient to induce EoE generation, the level of eosinophil, and incorporation of 5′-bromodeoxyuridine are both detected using immunohistochemical staining after intratracheal delivery of IL-13 into wild-type, STAT6, eotaxin-1, or IL-5-deficient mice. The result showed that the intratracheal delivery of IL-13 induced dose-dependent eosinophil accumulation in the esophagus and esophageal epithelial hyperplasia, and the inducing capability of IL-13 on EoE generation depends on the level of STAT6, eotaxin-1, and IL-5 (Mishra & Rothenberg, 2003). Subsequently, using inducible lung-specific IL-13 transgenic mice, EoE and esophageal tissue remodeling with epithelial hyperplasia are both induced; esophageal collagen, angiogenesis, and circumference of the esophagus are also affected after the overexpression of IL-13 in pulmonary tissue. In addition, transcriptomes between human EoE and IL-13-induced murine EoE are analyzed, and the result showed that 283 genes in IL-13-induced murine EoE overlapped with human EoE transcriptome, suggesting that these genes have conserved roles in experimental EoE and human EoE. Furthermore, EoE induced by IL-13 is dependent on eotaxin-1; however, tissue remodeling is independent on eosinophils and it can be inhibited by IL-13R alpha2 (Zuo et al., 2010).
After overexpression of IL-5 driven by the squamous epithelial promoter Epstein–Barr virus ED-L2 in the resident squamous esophageal epithelia of oxazolone haptenation-challenged L2-IL5 mice, dose-dependent panesophageal eosinophilia developed and included formation and degranulation of eosinophil microabscess and basal cell hyperplasia. Furthermore, the esophagus expressed increased levels of IL-13 and eotaxin-1. When corticosteroids were administered in these mice, the eosinophilia and epithelial inflammation were significantly reduced (Masterson et al., 2014). Another study revealed that omeprazole, a type of proton pump inhibitor, can block chemokine production independent of their inhibitory effects on gastric acid secretion. When human primary and telomerase-immortalized esophageal squamous cells are exposed to acidic bile salt medium, expression of IL-8 in these cells caused by the binding of nuclear factor-kappa B and activator protein-1 to IL-8 promoter is confirmed. Whereas, omeprazole in medium can not only inhibit the nuclear translocation of p65 and the binding of p65, c-jun, and c-fos to IL-8 promoter, thereby inhibiting the binding of nuclear factor-kappa B and activator protein-1 to IL-8 promoter and acidic bile salts-stimulated IL-8 expression, but also blocks the migration of immune cells induced by acidic bile salts (Huo et al., 2014). Our recent study found that BMP signaling was specifically activated in differentiated squamous epithelium; however, basal progenitor cells express follistatin, a BMP antagonist that could bind to BMPs (e.g., BMP4 and BMP7) and block their engagement with the receptors, and basal progenitor cell-specific expression of constitutively active BMP promoted squamous differentiation. Finally, we propose a model in which normal squamous differentiation of basal progenitor cells is mediated by BMP-driven NRF2 activation and basal cell hyperplasia is promoted by disruption of BMP signaling in EoE after validating in both a mouse EoE model and human biopsies (Jiang et al., 2015).
Tissue remodeling in EoE includes epithelial hyperplasia, subepithelial fibrosis, and hypertrophy of esophageal smooth muscle, and it can eventually lead to esophageal rings and strictures and mucosal fragility. Emerging studies revealed that tissue remodeling is caused by secretory products of eosinophils and mast cells, and the cytokines generated from surrounding esophageal cells, such as IL-4, IL-5, IL-13, eotaxin-3, periostin, and transforming growth factor-β1. All of these products and cytokines contribute to the process of tissue remodeling because of their profibrotic and remodeling effects or roles in regulating relevant proteins (Cheng, Souza, & Spechler, 2012). Therefore, inflammatory cytokines and basal cells closely correlate with EoE disease, and the roles of inflammatory cytokines in basal cells abnormalities and tissue remodeling of EoE are summarized in Figure 3. Animal models that simulate EoE remodeling have been well investigated using respiratory allergen-induced mouse (Mishra, Hogan, Brandt, & Rothenberg, 2001), IL-5 transgenic mouse (Mishra et al., 2008), IL-13 transgenic mouse (Zuo et al., 2010), and ovalbumin-challenged mouse (Rubinstein et al., 2011); however, to what extent these mouse models recapitulate the human EoE remains unknown.
Previously, the goal of EoE treatments mainly depends on decreasing the number of esophageal eosinophils, and measures for its treatments include the application of proton pump inhibitors, dietary modification, systemic and topical corticosteroids, and endoscopic treatments; all these methods have been extensively reviewed (Redd & Schey, 2013). Although cytokines participate in the process of EoE, few measures exist for specific targeting against cytokines. Thus far, anti-IL-5 antibody has been used in clinical trials for EoE therapy (Stein et al., 2006; Spergel et al., 2012); however, the efficiency of delivering antibody into cells needs to be improved because of their large molecular weight.
7 ROLES OF BASAL CELLS IN THE EAC FORMATION
As a leading cause of cancer deaths in developing countries, esophageal cancer mainly contains two types, EAC and ESCC, and tumorigenesis in the esophagus closely correlates with multiple potential risk factors, including the local dietary and cultural practices (Jakszyn et al., 2013; O'Doherty, Cantwell, Murray, Anderson, & Abnet, 2011; Sharp, Carsin, Cantwell, Anderson, & Murray, 2013), the consumption of tobacco (Cook et al., 2012; Hardikar et al., 2013), overweight and obesity (Kong et al., 2011; Turati, Tramacere, La Vecchia, & Negri, 2013), age and sex (Rubenstein, Scheiman, Sadeghi, Whiteman, & Inadomi, 2011), mutations in multiple genes such as p53 (Casson et al., 1994; Casson et al., 1998; Hollstein, Metcalf, Welsh, Montesano, & Harris, 1990), ras (Casson et al., 1997), NOTCH1 (Agrawal et al., 2012), PIK3CA (Phillips et al., 2006) and NRF2 (Shibata et al., 2011), and polymorphism in epidermal growth factor (EGF) (Menke et al., 2012) and matrix metalloproteinase genes (Cheung et al., 2012). Additionally, many signal transduction pathways are also involved in esophageal cancer progression (Babar et al., 2012; Ge et al., 2013; Liu et al., 2008; Pan et al., 2014; Yu et al., 2009; Zhang et al., 2012).
EAC is a minor type of malignant cancer whose frequency has dramatically increased, and the occurrence of EAC can be caused by erosive reflux disease (Erichsen et al., 2012), decreased estrogen exposure (Mathieu, Kanarek, Tsai, Rudin, & Brock, 2014), Helicobacter pylori infection (Liu, Wang, Wang, Li, & Gao, 2011), and single-nucleotide polymorphisms in cancer-related genes (Wu et al., 2011). Additionally, progress of EAC can also be further affected by Helicobacter pylori infection (Kountouras et al., 2012), methylation (Kaz et al., 2011; Wu, Bhagat et al., 2013a), and ectopic expression of some genes including GATA6 (Lin et al., 2012) and CDK6 (Ismail et al., 2011). As a precursor of EAC, Barrett esophagus can be promoted by inflammation and driven by the metaplasia process, this complicated process also involves many molecules including IL-1β, IL-5, IL-6, IL-13, and CDX. To trace the molecular and cellular etiology of EAC, several potential mechanisms have been proposed. The occurrence of EAC is often a sequential result of GERD-CDX expression-metaplasia-dysplasia, or it originates from the migration of Krt-7 positive embryonic cells to Krt-6 positive squamous cells or expansion and migration of gastric cardia progenitor cells induced by altered microenvironment at the SCJ.
Interestingly, other studies showed that obesity can also promote reflux and result in chronic inflammation and Barrett esophagus, thereby predisposing to adenocarcinoma (Kong et al., 2011; Ryan et al., 2011; Turati et al., 2013). To study the associations between obesity and EAC, 51 asymptomatic volunteers are divided into two groups, and the result showed the group with a large waist circumference exhibits significant lengthening of cardiac mucosa and shortening of the distal component of the LES, thereby leading to increased intrasphincteric acid reflux, damage to the SCJ, and expansion of adjacent cardiac glands. Moreover, the lengthening of the cardiac mucosa may arise from the metaplasia of the most distal esophageal squamous epithelium; therefore, a large waist circumference/obesity may be used to predispose the expansion of cardiac glands associated with EAC (Robertson et al., 2013). However, another group supports the viewpoint that the expansion of the gastric cardia originates from the proximal extension of original cardiac mucosa but not from the metaplasia of the most distal esophageal squamous epithelium (Quante, Abrams, & Wang, 2013).
Taken together, to decrease the mortality of EAC, preventive strategies including antireflux therapy, weight loss, and chemoprevention with nonsteroidal anti-inflammatory drugs and statins should be of key interest in both the primary and secondary prevention of EAC (Lagergren & Lagergren, 2013).
8 ROLES OF BASAL CELLS IN THE ESCC FORMATION
ESCC is the predominant type of esophageal cancer, and approximately 90% of esophageal cancer is ESCC in China. Currently, basal cells hyperplasia and dysplasia are both highlighted in the ESCC generation. Certainly, despite the presence of oncogenic lesions, cells in the state of terminal differentiation have relatively little effect on the whole tissue because of their rapid loss. However, because stem cells are permanent tissue residents, they have high potential to develop further deleterious changes that can induce tumor formation.
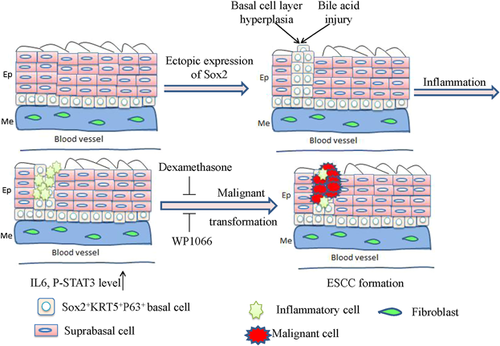
As a key transcription factor, Sox2 initially plays a role in the development and maintenance of undifferentiated embryonic stem cells, whereas emerging studies revealed that the amplification of SOX2 gene occurs in many types of cancer (Bass et al., 2009; Hussenet et al., 2010; McCaughan et al., 2010; Rudin et al., 2012); moreover, Sox2 is closely associated with clinical outcome and it also contributes to multiple malignant processes of cancer cells (Liu, Lin et al., 2013b; Liu et al., 2017). In our previous study, ectopic expression of Sox2 can cooperate with phosphorylated Stat3 to transform the foregut basal progenitor cells (Liu, Jiang et al., 2013a). We found that a population of basal progenitor cells, which have positive expression of Sox2 and Keratin5, has the potential of self-renewal and differentiation in vitro and in vivo. Although the ectopic expression of Sox2 is achieved in both forestomach and the esophagus, tumors can only be observed in the forestomach but not in the esophagus. Therefore, we hypothesized that inflammation in the forestomach may play a cooperative role in tumorigenesis. After administration of immunosuppressant dexamethasone, the tumor formation in the forestomach is dramatically reduced although Sox2 overexpression still occurs, and the same impact is also achieved with Stat3 inhibitor WP1066. These data suggest that the inflammation mediated by IL-1β/IL-6/Stat3 can cooperate with Sox2 overexpression to induce tumorigenesis in the forestomach although the esophagus and forestomach have the same structural layers. However, the downregulation of Sox2 and Stat3 in mouse and human squamous cancer cells reduce the growth of these cancer cells. In addition, the increased levels of phosphorylated STAT3 and Sox2 protein closely correlate with a poor prognosis for human ESCC (Liu, Jiang et al., 2013a), and this study reveals that the basal cells play a pivotal role in ESCC formation and are the origin of esophageal malignant transformation (Figure 4).
9 CONCLUSIONS
As we now know, mouse esophagus consists of multiple layers of squamous epithelial tissue and it is protected by sphincter muscle, the esophagus and forestomach are both lined with a similar stratified keratinized epithelium, and the forestomach is in direct continuity with the hindstomach. Although human esophagus is also protected by sphincter muscle, however, no keratinized squamous epithelium resides in human forestomach; thus, it is noticeable that the morphologic and physiological difference between mouse and human esophagus, implying the etiology and mechanism of esophageal diseases between mouse model and human may be somewhat different.
Because basal cells play pivotal roles in esophagus development, abnormalities in basal cells contribute to multiple esophageal diseases. A question is whether an exclusive subpopulation in basal progenitor cells exists for the specific esophageal disease or not? For example, does Barrett stem cell exist in esophagus? As we now know, cancer stem cells may originate from normal tissue or progenitor cells; thus, it may be intrinsic in the primary tumors at the early stage of tumorigenesis. Additionally, cancer stem cells may also derive from epithelial to mesenchymal transition caused by a creative microenvironment (Chaffer & Weinberg, 2011). Similar to cancer stem cells, the presence of Barrett stem cells that can maintain the Barrett metaplasia and their origin also remains underdetermined, and what causes the transformation of Barrett stem cells also remains to be determined. If it is true that each type of exclusive subpopulation in basal progenitor cells is responsible for its specific esophageal disease, another question is which factors induce the directed differentiation of the basal cells from the stem cell group? To dissect these mechanisms, a comprehensive comparison of gene expression profiles between transit amplifying cells in the PBL and stem cells in the IBL may be helpful to determine these factors or signal molecules involved in the direct differentiation and movement.
The progeny of esophageal stem cells may also exhibit different cell surface phenotypes at sequential stages of differentiation, another question is how to identify these exclusive subpopulations of basal progenitor cells and isolate them from esophagus? We think the most important thing is to identify their respective surface markers, as long as these cell surface markers are identified, the detailed process of cell differentiation in esophagus could be fully elucidated at the molecular and genetic level. Based on the identifications on cell surface markers, we can also screen imaging materials or molecules, which can specifically bind to the corresponding basal cells, to monitor and trace these basal cells during the pathological process.
Previous evidence has demonstrated that the alterations of DNA methylation and microRNA expression are both involved in the pathogenesis of esophageal diseases (Sun et al., 2013; Wu, Ajani et al., 2013b; Yu et al., 2014; Zhang et al., 2014); therefore, of most interest is exploration of the roles of these complicated alterations in esophageal basal cells during the pathogenesis of esophageal diseases, especially in metaplasia and cancer progression. Therefore, on one hand, isolating malignant basal cells at pivotal stages from animal diseases model and making comparisons with normal basal cells may be effective measures to understand this exquisite transformation process. On the other hand, development of imaging tools or reagents that can specifically identify basal cells will be helpful to monitor the real-time transformation and migration of basal cells during the pathological process.
Conventional therapeutic measures for esophageal cancer include surgery, radiotherapy, and chemotherapy, which have improved survival rates of patients. However, recurrence is possible and novel biotherapy measures, which target against pivotal molecules or malignant basal cells responsible for tumor formation, should be developed and attempted. As validated previously, both ESCC and EAC are caused by malignant basal cells driven by multiple factors and microenvironment change; thus, exploring the roles of these key molecules in transformation and removing these malignant basal cells/progenitor cells from niche may be a promising biotherapy measure for esophageal cancer therapy. To prevent these esophageal diseases, early detection when they are at a more curable stage is another potential therapeutic strategy. Therefore, exploring new biomarkers and measures for diagnosis is useful to evaluate the risks and stage of these diseases, and it is also helpful for therapeutic measures determination. Additionally, EoE, GERD, Barrett esophagus, EAC, and ESCC closely correlate with the inflammation process for responding esophagus damages; and avoiding damage caused by irritative dietary choices and the consumption of tobacco or alcohol may be other effective approaches to prevent the advent of these diseases.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (81302068 to KCL), the National High Technology Research and Development Program of China (2014AA020541 to KCL), the Program for the Top Young Innovative Talents of Fujian Province (to KCL), the International Collaborative Project of Fujian Province (2017I0014 to KCL), the Program of Fuzhou General Hospital for Distinguished Young Scientists Development (2016Q03 to FAX).
CONFLICTS OF INTEREST
All the authors declare no conflict of interest.
AUTHORS' CONTRIBUTIONS
LBS and XFA made substantial contributions to conception, design and collection of data. XZW, HXQ, and TLM searched the literature, edited the manuscript and revised English draft. LKC made substantial contributions and reviewed critically for important intellectual content and made substantial data contributions. All authors read and approved the final manuscript.




