Fluorescent multiplexing of 3D spheroids: Analysis of biomarkers using automated immunohistochemistry staining platform and multispectral imaging
Srabani Bhaumik and Jean Boyer are co-first authors.
Abstract
In preclinical cancer studies, three-dimensional (3D) cell spheroids and aggregates are preferred over monolayer cell cultures due to their architectural and functional similarity to solid tumors. We performed a proof-of-concept study to generate physiologically relevant and predictive preclinical models using non–small cell lung adenocarcinoma, and colon and colorectal adenocarcinoma cell line-derived 3D spheroids and aggregates. Distinct panels were designed to determine the expression profiles of frequently studied biomarkers of the two cancer subtypes. The lung adenocarcinoma panel included ALK, EGFR, TTF-1, and CK7 biomarkers, and the colon and colorectal adenocarcinoma panel included BRAF V600E, MSH2, MSH6, and CK20. Recent advances in immunofluorescence (IF) multiplexing and imaging technology enable simultaneous detection and quantification of multiple biomarkers on a single slide. In this study, we performed IF staining of multiple biomarkers per section on formalin-fixed paraffin-embedded 3D spheroids and aggregates. We optimized protocol parameters for automated IF and demonstrated staining concordance with automated chromogenic immunohistochemistry performed with validated protocols. Next, post-acquisition spectral unmixing of the captured fluorescent signals were utilized to delineate four differently stained biomarkers within a single multiplex IF image, followed by automated quantification of the expressed markers. This workflow has the potential to be adapted to preclinical high-throughput screening and drug efficacy studies utilizing 3D spheroids from cancer cell lines and patient-derived organoids. The process allows for cost, time, and resource savings through concurrent staining of several biomarkers on a single slide, the ability to study the interactions of multiple expressed proteins within a single region of interest, and enable quantitative assessment of biomarkers in cancer cells.
1 INTRODUCTION
The likelihood of phase I clinical trial investigational drug candidate to obtain Food and Drug Administration approval is a mere 5.1% in the oncology disease area, and attributed to lack of efficacy2 and unacceptable toxicity.3 Deficiencies in predictive preclinical cancer screening models contribute to high attrition rates. In the preclinical stages, many cancer drug screening and efficacy studies utilize conventional in vitro two-dimensional (2D) cell culture models that are basic and uncomplicated, but a poor representation of the physiological tumor environment. Compared to solid tumors, limitations of monolayer 2D cultures comprise of anchorage-dependent growth on flat, rigid, and usually synthetic surfaces, restricted cell-cell interactions, and forced cell polarity. Also absent are cell-extracellular matrix (ECM) interactions and oxygen-nutrient gradients, all of which affect cell metabolism, morphology, motility, growth, differentiation, invasion, cell-cycle kinetics, gene expression, cell function, and response to physiological stimuli such as drugs.
Several studies indicate that the use of 3D culture systems may mitigate drawbacks. Spheroids from 3D cultures recapitulate spatial and temporal configuration of in vivo tissues and are of particular interest in cancer research due to their similarity with solid tumors, such as apical polarity and gene expression profiles,4, 5 and cellular heterogeneity in patient-derived organoids. The multicellular and multilayered architecture of 3D spheroids consist of hypoxic, sometimes apoptotic and nonproliferating cells buried deep at the core of the structure, and proliferating cells on the outer surface, thereby creating a physiologically relevant tumor environment.6 Increased cell-cell interactions, internal zones with oxygen-nutrient gradients, different proliferation rates within the zones, and unequal drug exposure to cells within the spheroid structures bear similarity to many solid tumors. Monolayer 2D cultures require serial passaging every few days for maintenance, thereby accumulating genetic and phenotypic variance with the rise in the number of passages. Three-dimensional spheroids, on the other hand, maybe grown long term in a single passage for the duration of a drug efficacy study7 and thereby considered a promising platform for high-throughput (HTS) or high-content screening (HCS) of anticancer drugs. In recent years, multicellular 3D spheroids generated from immortalized cancer cell lines8 and organoids from patient-derived explants9 are routinely used to study in vitro preclinical cancer disease models10 and toxicity screening after drug treatment.11 In addition, 3D spheroids are utilized for studying newer anticancer drugs that address tumor heterogeneity,12 therapeutic resistance,13 and metastasis.14 Cancer drug sensitivity studies indicate higher chemoresistance in 3D spheroids derived from established prostate cancer,15 endometrial cancer,16 colon cancer,17 epithelial ovarian cancer cell lines,18 and patient-derived endometrial cancer19 compared to 2D cultures. Reviews of patient-derived 3D tumor models illustrate the advances in 3D spheroid technology towards improved drug development and preclinical studies.20, 21
Until recent years, pathology-based detection dominated the cancer diagnostics landscape, with immunohistochemistry (IHC) of tumor tissues as the hallmark for defining cancer types and subtypes. Automated chromogenic IHC assays validated for clinical tissue samples22-26 evaluate a single biomarker per slide but are ineffective in capturing the complex interrelationship between a multitude of other proteins in the cellular microenvironment. In the current and future diagnostic environment, studying the localization, spatial context, expression patterns, interaction between tumor and immune cells, and between endothelial and stromal cells in the tumor microenvironment is a necessity towards stratifying patients for cancer therapy. One of the limiting factors facing cancer tissue diagnostics is the scarcity of quality sample tissues. With the advent of molecular profiling of cancer and the ever-expanding spectrum of molecular diagnostic methods including microarrays, in situ hybridization, next-generation sequencing, and polymerase chain reaction-based technologies that utilize tissue as the starting material, demands on the limited supply of high-quality tumor tissue samples have increased several folds. Recent advances in multiplex IHC or immunofluorescence (IF) technologies effectively address this challenge by interrogating panels of multiple biomarkers on a single tissue section.27-29 In practice, however, chromogenic IHC multiplexing may be limited to three target biomarkers30 due to visualization and quantification constraints. A major drawback of chromogenic staining is nonlinear signal amplification and signal intensity saturation that renders the staining nonquantifiable. Chromogenic multiplexes are also difficult to visualize or assess due to signal overlap in instances where multiple biomarker proteins express in the same cellular compartment. In the fluorescent multiplex method, excitation and photoemission of specific wavelengths dictate the signal intensity, which is linear, directly proportional to target concentration, and quantifiable. Fluorescent IF multiplexing allows for simultaneous detection of a far greater number of targets, ranging from 5 or more biomarkers31, 32 to 61 protein epitopes.33 The fluorescent method thus allows for more information from sparse tissue samples such as core needle biopsies, small metastatic, and rare tumors in the spatial context of cells, and allows visualization of colocalized biomarkers. The option to visualize individual signals stored in separate channels is an advantage of fluorescent multiplex staining. By turning on only the fluorescent channel of interest, one can visualize uniplex signals of a multiplex scan. The spectral overlap is a challenge in both methods, but in fluorescent multiplexing, autofluorescence removal and signal separation through spectral unmixing34 allow for increased sensitivity, improved specificity, and accurate signal discrimination by minimization of fluorophore crosstalk.
In this study, we describe a proof-of-concept workflow to generate preclinical models of colorectal and lung cancer based on IF multiplexing of formalin-fixed paraffin-embeddied (FFPE) sections from cell line-derived 3D spheroids and aggregates. A prerequisite of biomarker-based cancer therapies includes IHC and molecular test-based screening for specific biomarkers prevalent in certain cancer types. We investigated expression patterns of key biomarkers routinely analyzed in non–small cell lung cancer (NSCLC), and colon and colorectal cancer (CRC) adenocarcinomas. In the NSCLC model, we examined proteins expressed from genes with targetable oncogenic driver mutations (ALK and epidermal growth factor receptor [EGFR]). In the CRC model, we examined proteins expressed from the oncogenic BRAF V600E gene and mismatch repair (MMR) genes MSH2 and MSH6. Determining Cytokeratin 7 (CK7) and CK20 protein expression allowed differentiation between lung adenocarcinoma (mostly CK7 positive) and colorectal adenocarcinoma (mostly CK20 positive, with high positivity in metastatic tumors).35, 36 We first performed chromogenic 3,3′-diaminobenzidine (DAB) uniplex automated IHC staining of individual biomarkers on 3D spheroid and aggregate sections using assay protocols validated for staining clinical tissues.22-24 Following the corroboration of uniplex IF and chromogenic patterns, multiplex slides were generated. We then utilized spectral unmixing of these multiple fluorescence signals to remove signal overlaps and autofluorescence and generate a distinct, precise signal for each color channel. The fluorescent signals were next quantitated by automated image analysis methods. We demonstrate the utility of a combined 3D spheroid and fluorescent multiplexing workflow with automated quantitation for advancing opportunities in preclinical drug testing and personalized cancer therapies.
2 MATERIALS AND METHODS
2.1 Culture of adenocarcinoma cell lines
Two non–small cell lung adenocarcinoma (NSCLC) cell lines NCI-H2228 and NCI-H1975, and two colon and CRC adenocarcinoma cell lines HT-29 and KM12 were used in this study. NCI-H2228 (non–small cell lung adenocarcinoma), NCI-H1975 (non–small cell lung adenocarcinoma), and HT-29 (colorectal adenocarcinoma) cell lines were acquired from American Type Culture Collection (ATCC, Manassas, VA). KM12 (colon adenocarcinoma) cell line was acquired from the NCI-Fredrick Cancer DCTD Tumor/Cell line Repository (NCI, Frederick, MD). Media (Roswell Park Memorial Institute Medium 1640 for NCI-H2228, NCI-H1975, and KM12 cells, and McCoy's 5A Medium Modified for HT-29 cells) were purchased from ATCC and supplemented with 1% penicillin-streptomycin (HyClone GE Healthcare Life Sciences, Logan, UT). Media for NCI-H2228, NCI-H1975, and HT-29 cells were supplemented with 10% Seradigm premium fetal bovine serum (VWR Life Science, Radnor, PA), and medium for KM12 cells were supplemented with 20% fetal bovine serum (Seradigm). Cells were cultured in 3D culture systems (see Section 2.2) and maintained according to manufacturers’ specifications. All cell lines were certified mycoplasma free.
2.2 Three-dimensional cell culture and FFPE
Two 3D culture systems, AlgiMatrix (alginate scaffold six-well plates; Cat. A10982-02) and the Nunclon Sphera (scaffold-free T75 flasks; Cat. 174952), were purchased from Thermo Fisher Scientific (Waltham, MA). HT-29 cells were seeded at 2 × 105 per well on Algimatrix plates and grown for 12 days for aggregate formation before harvesting. NCI-H2228, NCI-H1975, and KM12 cells were seeded at 5 to 6 × 106 per Nunclon Sphera T75 flask and grown for 4 days for spheroid formation before harvesting. Spheroids and aggregates were processed in 10% neutral-buffered formalin and then encapsulated in Richard-Allen Scientific HistoGel Specimen Processing Gel (Thermo Fisher Scientific; Cat. HG-4000-012). HistoGel-encapsulated samples were fixed overnight in Excelsior ES Tissue Processor (Thermo Fisher Scientific) followed by paraffin embedding on a Sakura Tissue-Tek TEC Console (VWR Scientific, Radnor).
2.3 Antibodies for chromogenic IHC and IF multiplex staining
We obtained primary, secondary, and tertiary antibodies for chromogenic IHC and IF multiplex staining from Roche Tissue Diagnostics, Tucson, AZ, unless otherwise noted. For staining lung adenocarcinoma 3D sections, the following primary antibodies were used: VENTANA ALK (D5F3) CDx Assay Rabbit Monoclonal (Cat. 790-4796), CONFIRM anti-EGFR (5B7) Rabbit Monoclonal Primary Antibody (Cat. 790-4347), anti-Thyroid Transcription Factor-1 (TTF-1) (SP141) Rabbit Monoclonal Primary Antibody (Cat. 790-4756), and CONFIRM anti-Cytokeratin 7 (SP52) Rabbit Monoclonal Primary Antibody (Cat. 790-4462). For staining colorectal adenocarcinoma 3D sections, the following primary antibodies were used: anti-BRAF V600E (VE1) Mouse Monoclonal Primary Antibody (Cat. 790-4855), anti-MSH2 (G219-1129) mouse monoclonal (Cat. 760-4265), CONFIRM anti-MSH6 (44) Mouse Monoclonal Primary Antibody (Cat. 790-4455), and CONFIRM anti-Cytokeratin 20 (SP33) Rabbit Monoclonal Primary Antibody (Cat. 790-4431). For both chromogenic IHC and IF multiplex staining, the secondary antibodies OmniMap anti-Rb HRP (RUO) DISCOVERY (Cat. 760-4311) and OmniMap anti-Ms HRP (RUO) DISCOVERY (Cat. 760-4310) were used.
2.4 Automated chromogenic IHC of FFPE 3D spheroid sections
Serial sections of 4 µm were cut from FFPE blocks containing 3D spheroids and aggregates and mounted on positively charged glass slides (Superfrost Plus, Cat. 4951PLUS4; Thermo Fisher Scientific). We performed all chromogenic IHC assays on the BenchMark ULTRA IHC/ISH platform (Roche Tissue Diagnostics) using VENTANA/Roche reagents (Roche Tissue Diagnostics, Tucson, AZ). Slides were deparaffinized using EZ-Prep (Cat. 950-100) followed by epitope retrieval with Cell Conditioning 1 buffer (Cat. 950-124 or Cat. 950-500). One serial section stained with secondary antibody alone served as a negative control. For chromogenic (DAB) staining, some slides were developed with the ultraView Universal DAB Detection Kit (Cat. 760-500) or OptiView DAB IHC Detection Kit (Cat. 760-700), with or without amplification with OptiView Amplification Kit (760-099), according to manufacturer's specifications. The biomarkers and antibodies for automated chromogenic DAB staining are listed in Table 1.
| Panel | Biomarker proteins | Primary antibody |
|---|---|---|
| NSCLC | EGFR | CONFIRM anti-EGFR (5B7) Rabbit Monoclonal Primary Antibody |
| NSCLC | TTF-1 | Anti-Thyroid Transcription Factor-1 (SP141) Rabbit Monoclonal Primary Antibody |
| NSCLC | ALK | Anti-ALK (D5F3) Rabbit Monoclonal Primary Antibody |
| NSCLC | CK7 | CONFIRM anti-Cytokeratin 7 (SP52) Rabbit Monoclonal Primary Antibody |
| CRC | BRAF V600E | Anti-BRAF V600E (VE1) Mouse Monoclonal Primary Antibody |
| CRC | MSH2 | MSH2 (G219-1129) Mouse Monoclonal Primary Antibody |
| CRC | MSH6 | CONFIRM anti-MSH6 (44) Mouse Monoclonal Primary Antibody |
| CRC | CK20 | CONFIRM anti-Cytokeratin 20 (SP33) Rabbit Monoclonal Primary Antibody |
- Abbreviations: CRC, colorectal cancer; DAB, 3,3′-diaminobenzidine; NSCLC, non–small cell lung cancer.
2.5 Automated IF multiplex staining process
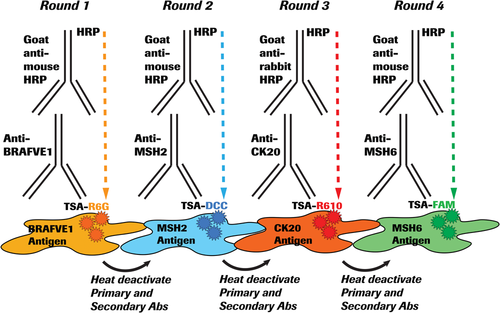
Fluorescent 4-plex staining of 3D spheroids and aggregates were performed using the U Discovery 5-Plex RUO staining procedure,37 which utilizes tyramide signal amplification (TSA).29, 38 The assay was run on the VENTANA DISCOVERY ULTRA automated staining platform (Roche Tissue Diagnostics) using VENTANA/Roche reagents, except where noted. The fully automated IF multiplex protocol involves deparaffinization (EZ-Prep solution; Cat. 950-102) and antigen retrieval (DISCOVERY CC1 solution; Cat. 950-500) at high temperature followed by four sequential rounds of (a) incubation of target antigen with a primary antibody, (b) binding of primary antibody to horseradish peroxidase (HRP)-conjugated antispecies (goat anti-mouse or goat anti-rabbit) secondary antibody, (c) enzymatic reaction of HRP in the primary-secondary antibody complex with the tyramide-fluorophore detection system, and (d) covalent binding of the tyramide-fluorophore complex to the tissue. A heat-deactivation step between each round of staining removed the unbound antibodies from the preceding cycle. A total of three heat-deactivation steps were needed for the 4-Plex assay. After the completion of four staining rounds, the sections were counterstained with quantum dot-4′,6-diamidino-2-phenylindole (QD-DAPI) (Cat. 760-4196). ProLong Diamond antifade mounting media (Cat. P36961; Thermo Fisher Scientific) was applied to the stained sections for coverslipping. The fluorescent detection kits (Roche Tissue Diagnostics), available in prefilled dispensers and with real-time stability of 18 months, are designed for use with the VENTANA DISCOVERY ULTRA system. The fluorescent detection kits used were DISCOVERY Rhodamine 6G Kit (Cat. 760-244), DISCOVERY DCC Kit (Cat. 760-240), DISCOVERY Red 610 Kit (Cat. 760-245), DISCOVERY FAM Kit (Cat. 760-243), and DISCOVERY Cy5 Kit (Cat. 760-238). The staining procedure was standardized across several reagent formulations, temperature ranges, durations of heat deactivation, and stability of the fluorophores as described.29, 37 The antibody incubation time and effect of heat deactivation were optimized for each biomarker before the final protocol design. The U Discovery 5-Plex RUO IF staining procedure has a locked fluorophore sequence in the following positions (a) rhodamine 6G (R6G), (b) DCC, (c) Red610, (d) Cy5, and (e) FAM. Two different automated staining protocols were designed for NSCLC and CRC. The biomarker-fluorophore pairing, staining parameters, and staining sequence in the protocols are discussed in Table 2.
| Panel | Biomarker protein | Primary antibody | Primary antibody incubation time, min | Secondary antibody incubation time, min | Fluorophore | TSA-fluor deposition time, min |
|---|---|---|---|---|---|---|
| NSCLC | EGFR | Anti-EGFR (5B7) | 16 | 12 | R6G | 8 |
| NSCLC | TTF-1 | Anti-TTF-1 (SP141) | 16 | 12 | Red610 | 8 |
| NSCLC | ALK | Anti-ALK (D5F3) | 20 | 12 | Cy5 | 8 |
| NSCLC | CK7 | Anti-Cytokeratin 7 (SP52) | 8 | 12 | FAM | 8 |
| CRC | BRAF V600E | Anti-BRAF V600E (VE1) | 16 | 12 | R6G | 8 |
| CRC | MSH2 | Anti-MSH2 (G219-1129) | 16 | 12 | DCC | 12 |
| CRC | CK20 | Anti-Cytokeratin 20 (SP33) | 16 | 12 | Red610 | 8 |
| CRC | MSH6 | Anti-MSH6 (44) | 16 | 12 | FAM | 8 |
- Abbreviations: CRC, colorectal cancer; IF, immunofluorescence; NSCLC, non–small cell lung cancer; TSA, tyramide signal amplification.
2.6 Fluorescent-labeled slide scanning and spectral unmixing
Whole-slide scans of the fluorescent-labeled sections were obtained at ×10 magnification using the Vectra 3.0 Imaging System (Akoya Biosciences, Menlo Park, CA). Multispectral images of the selected fields of view (FOV) were scanned at ×20. The multispectral camera in the scanner acquired images at every 20-nm interval ranging from 440 to 720 nm. A spectral library was constructed in the inForm 2.4 Image Analysis Software (Akoya Biosciences) by spectrally analyzing each antibody-fluorophore pair according to the manufacturer's recommendation. The spectral library was designed for DAPI (460 ± 25 nm), DCC (480 ± 15 nm), FAM (520 ± 10 nm), R6G (572 ± 10 nm), Red 610 (625 ± 15 nm), and Cy5 (690 ± 25 nm) emission spectra. Spectral signal extracted from each fluorophore was saved in the spectral library. Autofluorescence was assigned the black channel while fluorophores were assigned the pseudocolors red (Red 610), green (FAM), cyan (DCC), magenta (Cy5), orange (R6G), and blue (DAPI). FOV images were spectrally unmixed using inForm 2.4 and then merged to obtain the final multiplex images. To obtain uniplex images, each spectrally unmixed channel was individually selected in addition to the DAPI channel.
2.7 Analysis of fluorescent multiplex images
Automated cell segmentation was performed with the inForm 2.4. All five FOVs and autofluorescence images were loaded onto the software. Segmentation was performed based on nuclear counterstain (DAPI). From the image configuration table, adaptive cell segmentation and score modules were selected for analysis. The cell segmentation setting included nucleus, cytoplasm, and membrane compartments. For optimum cell segmentation, biomarkers that stained one of the nuclear, membranous, or cytoplasmic compartments were selected. Auto-adaptive threshold was used to establish fluorescent intensity cutoff before cell segmentation. The parameters were then tested and adjusted to obtain the best segmentation using feedback from the live preview window. The final settings were applied to all the FOV images and the segmentation output captured.
In the cell segmentation data module, the pixel intensity of each biomarker and their associated fluorophore were extracted from relevant cellular compartments. Mean biomarker pixel intensities of 500 cells (with intensities above the mean threshold) were next analyzed to obtain average value and standard deviation of the mean (n = 4-5).
The colocalization algorithm of inForm 2.4 was used to score multipositive cells with coregistered biomarkers. The percentages of the double-positive nuclear stain of MSH2-DCC and MSH6-FAM in the 3D CRC cells were determined. In the double-positivity scoring module for each marker setting, the fluorophore component and threshold maximum in the nuclei compartment were selected to quantify percent positive and negative cells in the FOV. The final output consisted of single-positive MSH2-DCC (cyan), single-positive MSH6-FAM (green), double-positive MSH6-FAM and MSH2-DCC (magenta), and biomarker-negative (gray) cells.
GraphPad Prism 8.0 was used to generate plots from the analytical output of inForm software. Violin plots were generated from the average value and standard deviation of the mean pixel intensity of each biomarker. Stacked column graphs representing a fraction of total were generated using the mean percentages of single-positive and double-positive cells in several FOV of KM12 and HT-29 cells.
3 RESULTS
3.1 Primary antibody selection and IF staining concept
In this proof-of-concept study, we utilized a panel of four key biomarkers per cancer indication to generate a workflow for the simultaneous detection of multiple biomarkers on a single slide. The cell lines for 3D included both expressors and nonexpressors of the biomarkers to establish positive- and negative-staining patterns. To capture a broad representation of biomarker expression, and to visualize the interplay between multiple biomarkers following IF multiplexing, we selected proteins whose cellular and subcellular expression locales ranged from membranous to cytoplasmic and nuclear. The NCI-H2228 and NCI-H1975 (NSCLC) slides were stained with ALK (cytoplasmic expression), EGFR (membranous to diffuse cytoplasmic expression), TTF-1 (nuclear expression), and CK7 (cytoplasmic expression) primary antibodies. The HT-29 and KM12 (CRC) slides were stained with BRAF V600E (VE1) (cytoplasmic expression), MSH2 (nuclear expression), MSH6 (nuclear expression), and CK20 (cytoplasmic expression) primary antibodies. Following primary and secondary antibody selection, chromogenic uniplex DAB staining of each biomarker was first performed using on-market staining protocols. Next, a 4-plex IF staining was performed on NSCLC and CRC slides with automated staining protocols developed from the U DISCOVERY 5-Plex IF staining procedure on the VENTANA DISCOVERY ULTRA platform.37 The TSA-based IF multiplexing involves the binding of unconjugated primary antibody to target antigens on the cells, followed by recognition of the primary antibody by HRP-conjugated antispecies secondary antibody. Next, in an enzymatic reaction, HRP catalyzes fluorescent conjugated tyramide (TSA-fluor) to generate activated tyramide. Multiple copies of activated tyramide form covalent bonds with tyrosine residues on or near the antigens and are deposited along with the associated fluorophores on the cells. Heat deactivation followed by washes removes the excess unbound antibodies and the primary-secondary antibody complexes while retaining the tyramide-fluorophore on the antigen. These steps are sequentially repeated in multiple rounds using different combinations of antibodies and fluorophores. Nuclear counterstain DAPI is applied after the final round. The removal of primary-secondary pairs after each cycle allows the use of multiple same-species primary antibodies without crosstalk concerns. The order of antibody-fluorophore pairing in the CRC panel is illustrated in Figure 1.
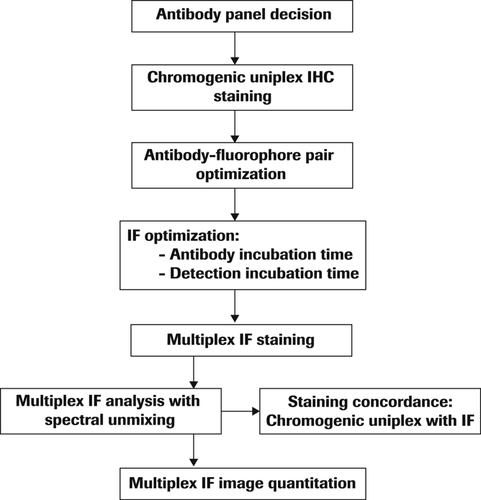
To create a balanced IF multiplexing protocol, antibody titration, incubation times, and removal of unstained antibody complex by heat deactivation followed by the number of washes between iterative cycles were optimized. The IF multiplexing protocol workflow is outlined in Figure 2.
3.2 Confirmation of biomarker protein expression by uniplex chromogenic IHC staining of 3D spheroids and aggregates
Each primary antibody in the NSCLC and CRC panels was optimized for chromogenic staining of FFPE 3D spheroid and aggregate sections on the BenchMark ULTRA platform following manufacturer's recommendations.
In the NSCLC panel (Figures 3A,B), ALK (D5F3) antibody contributed to cytoplasmic and diffused cytoplasmic staining patterns in 3D spheroid sections of the lung adenocarcinoma cell line NCI-H2228 but showed lack of staining in 3D spheroids of NCI-H1975. Strong membranous to diffuse cytoplasmic EGFR staining was observed in both NCI-H2228 and NCI-H1975 with the EGFR (5B7) antibody. CK7 (SP52) and TTF-1 (SP141), biomarkers used to distinguish lung adenocarcinomas from squamous cell carcinomas,39 demonstrated strong cytoplasmic and nuclear expression, respectively, in adenocarcinomas NCI-H2228 and NCI-H1975.
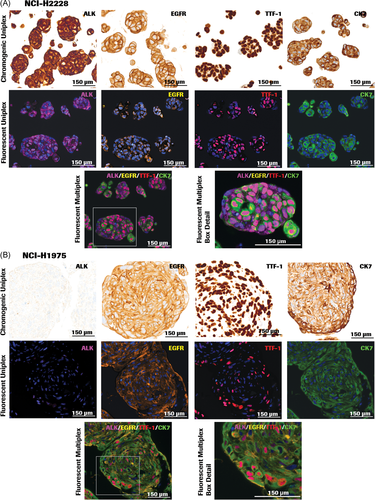
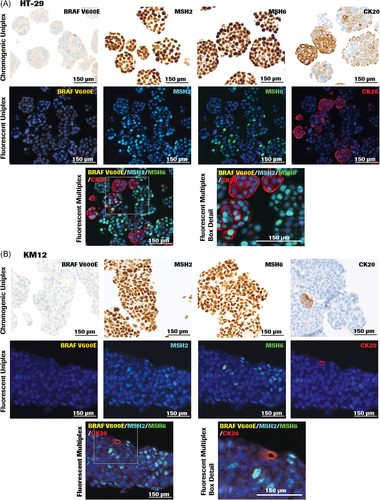
In the CRC panel containing sections from 3D colon adenocarcinoma (KM12) spheroids (KM12) and colorectal adenocarcinoma (HT-29) aggregates (Figure 4A,B), BRAFV 600E (VE1) antibody demonstrated cytoplasmic staining in HT-29 cell line, which harbors the BRAF V600E gene mutation. The antibody shows the absence of staining in KM12 cells, which contain wild-type BRAF, corroborating published data.40 MSH2 (G219-1129) and MSH6 (44) antibodies stained the MMR proteins in both KM12 spheroids and HT-29 aggregates as expected, since both cell lines are MMR proficient.
3.3 Adjusting protocol parameters to standardize IF staining
For chromogenic IHC staining, we used on-market primary antibody dispensers and validated each marker using the appropriate DAB Detection Kit. We used the same dispensers for fluorescent staining, thereby keeping the concentration of primary antibodies identical between the two procedures. Serial dilution (titration) of the primary antibody or decreasing the antibody incubation time is recommended for high-expressing proteins to minimize signal bleed-through across spectrally adjacent channels. For a high-abundance protein such as CK7 in the NSCLC panel, adjusting the primary antibody incubation time from 12 to 8 minutes markedly reduced the signal bleed-through to adjacent channels (Table 2).
3.4 Select antibody-fluorophore pair and determine multiplex staining order
Optimization of IF staining is dependent on both staining and imaging parameters. In many instances, the fluorescent signals from antibodies that detect high-abundance proteins may cause bleed-through and obscure low-intensity biomarker signals in adjacent channels. One way to mitigate the challenge is balancing signals across all four fluorophore channels by titration of high-expressing proteins, modifying antibody incubation times, and pairing with a fluorophore that results in an optimal signal. Choosing the sequence of biomarkers expressing in different compartments or locales within the cells is also taken into consideration. In addition, each epitope is tested to withstand multiple rounds of heat deactivation in the staining procedure. In the NSCLC panel, the CK7 antibody was paired with the fluorophore FAM and placed at the last round of staining sequence, because the CK7 epitope is capable of withstanding three rounds of heat deactivation. The CK7-FAM pair maintained robust signal intensity, as shown in Figure 3A,B.
In the CRC panel (Figure 4A,B), the primary antibody to detect low-signal protein BRAF V600E was paired with the fluorophore R6G and placed in the first round of the staining sequence, which was not subject to a heat-deactivation cycle. The R6G-BRAF V600E (VE1) pair retained optimal signal intensity in the CRC panel, while the R6G-EGFR pairing was optimum for the NSCLC panel.
3.5 Signal balance across fluorophore channels
Before imaging multiplex signals, it is important to ensure that the signals for individual biomarkers fall within an equivalent dynamic range. Adjustment to protocol parameters is necessary to obtain balanced signals with minimal crosstalk. For these studies, we modified the protocol parameters as needed by adjusting the incubation time of primary antibodies or the time for TSA-fluorophore deposition. Protocol parameters for fluorescent multiplexing are shown in Table 2. For MSH2 (CRC panel) increasing the TSA-DCC deposition time from 8 to 12 minutes generated optimal signal intensity with minimal crosstalk. In the NSCLC panel, primary antibody incubation times were modified to 8 minutes for CK7 and 20 minutes for ALK to optimize the signals.
In instances where expression locales of multiple biomarkers overlap, it is important to balance the channels such that overlapping signals are identified as colocalized in a multiplex IF. This involves the judicious selection of each antibody-fluorophore pair during the multiplex protocol design. Meticulous titration of each pair is required for multiplex IF to ensure that during colocalization, the signal from one channel does not obscure the overlaid signal from the second channel. In addition, the color of the colocalized signals should be identifiable as a blend of the two (or more) pseudocolors. In the CRC panel, nuclear proteins MSH2 (DCC: cyan) and MSH6 (FAM: green) colocalized on several nuclei to impart a blue-green color that was easy to distinguish from the distinct cyan and green signals of singly-stained nuclei (Figure 4A,B).
3.6 Detect biomarker expression by multiplex IF
We studied the expression of NSCLC or CRC biomarkers on single FFPE slides of 3D spheroids and aggregates stained by multiplex IF. In all slides, the nucleus was defined by DAPI counterstain. A 4-plex IF staining protocol optimized for the NSCLC panel was applied to FFPE sections of NCI-H2228 and NCI-H1975 3D spheroids (Figure 3A,B). The biomarker-fluorophore pairs were EGFR-R6G, TTF-1-Red610, ALK-Cy5, and CK7-FAM. The pseudocolors assigned for fluorescence detection were orange (R6G), red (Red610), green (FAM), and magenta (Cy5). In both NCI-H2228 and NCI-H1975 3D sections, TTF-1-Red610 displayed strong nuclear signal in red, and CK7-FAM exhibited strong cytoplasmic signal in green. A membranous and diffuse cytoplasmic stain was observed with EGFR-R6G (orange) in both cell lines. ALK-Cy5 (magenta) depicted strong cytoplasmic staining in NCI-H2228 (Figure 3A), but negligible staining in NCI-H1975 (Figure 3B).
A 4-plex IF staining protocol designed for the CRC panel was applied to FFPE sections of HT-29 and KM12 3D cell aggregates and spheroids (Figures 4A,B). The biomarker-fluorophore pairs used were BRAF V600E-R6G, MSH2-DCC, CK20-Red610, and MSH6-FAM. The pseudocolors assigned for fluorescence detection were cyan (DCC), green (FAM), orange (R6G), and red (Red610). MSH2-DCC (cyan) and MSH6-FAM (green) stained the nuclei either singly or as a combination. Colocalization of MSH2 and MSH6 in the nuclear compartment was depicted by merged staining of a blue-green color. Strong cytoplasmic staining in red was observed with CK20-Red610. HT-29 sections showed robust CK20-Red610 staining compared to KM12. BRAF V600E-R6G (orange) displayed weak cytoplasmic staining in HT-29 (Figure 4A) and negligible staining in KM12 sections (Figure 4B).
One of the challenges of fluorescent multiplexing is crosstalk between neighboring channels due to the broad emission spectrum of several of the fluorophores. The autofluorescence and signal overlap in IF stained slides were mitigated through spectral unmixing using the inForm 2.4 software, resulting in multiplex IF images with clearly delineated signals from each antibody-fluorophore pair (Figures 3A,B and 4A,B, bottom rows), and identifiable colocalized signals (Figure 4A,B, bottom rows).
3.7 Chromogenic IHC and IF staining pattern concordance
We tested the staining pattern concordance to ensure that the performance of IF multiplexing was consistent with the on-market DAB chromogenic IHC. Spectrally unmixed single-marker (uniplex) IF images representing each biomarker (Figures 3A,B and 4A,B middle rows) were generated from the multiplex composite images (Figures 3A,B and 4A,B bottom rows). Each biomarker-fluorophore channel was selected along with the DAPI channel to generate the series of uniplex images (Figures 3A,B and 4A,B middle rows) while the rest of the channels were turned off. Visual comparison of uniplex chromogenic IHC and uniplex IF staining patterns indicated that IF pattern of each biomarker was specific and matched the chromogenic staining pattern and distribution (Figures 3A,B and 4A,B, top and middle rows).
3.8 Image analysis of multiplex IF
Images from the multiplex IF of both NSCLC and CRC panels were analyzed by DAPI-dependent cell segmentation and intensity measurement using the inForm 2.4 software. In a multiplex IF, the heat-deactivation step between each round of biomarker staining could affect downstream antigens and fluorophores, resulting in either reduced or increased signal intensities in the later rounds. The sequence of antibody-fluorophore pairs in the multiplex scheme may also affect signal intensity. Therefore, we did not perform signal intensity-based comparison between different biomarkers within each cell line. We also did not correlate biomarker signal intensities with their expression levels but assessed the mean signal intensity of the same antibody-fluorophore pair (intra-biomarker assessment) relative to a threshold value to determine the positive or negative expression of a biomarker. Evaluation of the same biomarker in both the cell lines within a panel determined the dynamic range of fluorescence intensities for a specific antibody-fluorophore pair since it was positioned in the same staining round of the IF multiplexing workflow. Adaptive cell segmentation (inForm 2.4), optimized to identify different cellular components, demarcated NSCLC biomarkers expressed in the nucleus, cytoplasm, and membrane compartments of a representative 3D NCI-H2228 spheroid section (Figure 5A). The mean signal intensities of 500 fluorescent-labeled cells from five FOVs per slide for each biomarker in the NSCLC and CRC panels were calculated to assess inter-cell line variability (Figure 5B-E). In the NSCLC panel, NCI-H2228 cells demonstrated expression of ALK (Figure 3A), while the nonexpressor cell line NCI-H1975 showed no positive signal from the biomarker, corroborating observations from the chromogenic DAB staining pattern (Figure 3B), and confirming negative expression of the biomarker. Mean nuclear signal intensity of TTF-1 was threefold higher in NCI-H2228 compared to NCI-H1975, whereas the signal intensity of EGFR was equivalent, although at low levels in both the cell lines. CK7 indicated high signal intensity in NCI-H2228 in comparison to NCI-H1975, the difference being four-fold higher in NCI-H2228 cells (Figure 5B,C).
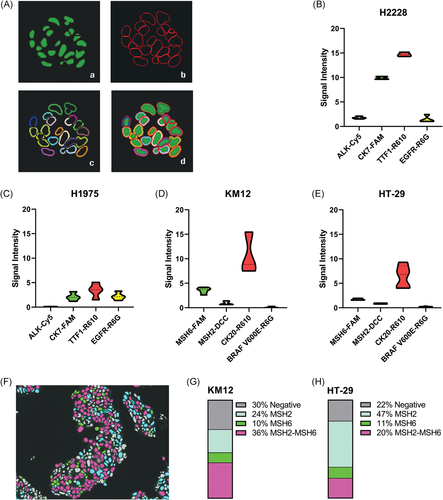
In the CRC panel, both KM12 and HT-29 cells showed equivalent signal intensities of MSH2, MSH6, and BRAF V600E, whereas CK20 signal intensity was higher in KM12 cells (Figure 5D,E). In these two cell lines, MSH2 and MSH6 colocalized in the nucleus of several cells, leading us to determine the percentage of cells containing either singly-expressed or coexpressed biomarkers (Figure 5F). Automated colocalization analysis of MSH2 and MSH6 in KM12 and HT-29 cells allowed quantitative evaluation of single-positive and double-positive cells in large sample sizes and multiple FOVs, highlighting different proportions of single- and double-positive biomarker expressions. In comparing the fraction of total analyses, KM12 cells indicated a higher percentage of double-positive (mean score of 36% vs 20%) and biomarker-negative (mean score of 30% vs 22%) cells relative to HT-29 (Figure 5G,H). In both cell lines, MSH6 was expressed singly in 10% to 11% of cells, whereas MSH2 was expressed singly in 24% of KM12 cells, in comparison to twice the percentage in HT-29 cells (47%) (Figure 5G,H).
4 DISCUSSION
This study describes the development and implementation of an automated IF multiplexing method for FFPE sections of 3D spheroids and aggregates to detect the concomitant expression of multiple key NSCLC and CRC biomarkers on a single slide. Contrary to many studies that utilize manual staining to study the expression of biomarkers, our method takes advantage of automated staining platform(s) and multispectral imaging (MSI) analysis for ease-of-use workflow combined with standardized procedures for reliable results. Our studies also indicate that IF multiplexing can substitute conventional uniplex chromogenic staining to generate maximum biomarker-specific information from a single section. We believe that our workflow design of multiplex IF using customizable antibody panels and software-aided image analysis makes a robust platform for preclinical cancer studies.
During the development of IHC and in situ hybridization-based diagnostic assays, FFPE slides prepared from 2D-cultured cell pellets of tumor or genetically engineered cell lines provide biologically relevant positive and negative controls. FFPE slides from 2D-cultured cells are adequate for these applications as the goal is visualization of the expression of a single biomarker, but there are no additional benefits of generating FFPE control slides from 3D cultured spheroids. Most in vitro preclinical testing methods, however, are developed in conventional 2D human cell cultures and xenografts. In preclinical drug screening and toxicity studies, such 2D-cultured cell models are proving to be increasingly unreliable as predictors of clinical efficacy and toxicity, as evidenced by massive drug failure rates in late clinical stages. Comparison of 2D and 3D breast cancer cells in drug sensitivity and resistance studies showed that 3D cells demonstrated lower cell viability, decreased drug efficacy, and increased activity of proteins involved in cell survival, drug export, and resistance,13 indicating overestimation of efficacy data from conventional 2D cultures. The failure of 2D cells in drug screening studies is attributed to the artificial cellular environment, which contributes to a lack of functional capabilities such as cell-cell interaction, cell-ECM signaling, and the tumor microenvironment. Three-dimensional-cultured cells, which partially mimic the in vivo cellular environment of tumors, mitigate most of these limitations.
The poor correlation between preclinical studies and clinical trials in cancer therapeutics is a driving factor for developing an IF multiplexing method using 3D spheroids. Devising rapid and efficient 3D spheroid models that recapitulate the pathophysiology of solid tumors is crucial for decreasing attrition rates and for the successful translation of cancer drugs to clinics. As the 3D spheroid technology becomes more cost-effective, simplified, and standardized, new and improved screening platforms are being developed to fulfill the escalating demand for preclinical drug discovery, and HTS and HCS drug efficacy studies. The ability to study the effect of drug treatment on multiple biomarkers at the same time is also a necessity for the future in vitro 3D models. The 3D spheroids in our studies, established in a standard cell culture laboratory and embedded in paraffin blocks using conventional IHC protocols,41 indicate easy adaption of the workflow in R&D laboratories.
In recent years, rapid strides in technologies focused on multiplexed IF of protein biomarkers in FFPE tissues of tumors and their microenvironment resulted in the development of diagnostic and predictive tools of patient stratification for immunotherapy. There are no known reports of multiplexing studies with FFPE 2D-cultured cells, possibly because of the limited benefits of this cellular model in drug screening and efficacy studies. In contrast, 3D spheroids from cancer cell lines or patient-derived cells, in combination with advanced multiplexing technologies, offer tremendous potential in preclinical high-throughput drug screening, efficacy, and toxicity studies. We leveraged our proficiency in IF multiplexing technology developed for FFPE tumor tissues37 and applied it to FFPE slides derived from 3D-cultured cancer cells because the biological complexity of 3D cells is closer to tumors. To generate a robust IF multiplexing assay with 3D cells as described in our study, it is imperative to have expertise in automated IHC and IF, fluorescent multiplexing of cancer tissues, multispectral imaging, and data analysis, as well as in generating 3D-cultured cells from cancer cell lines. Our in-house capabilities in all of the above enabled us to develop a workflow that encompasses all aspects of IF multiplexing, with 3D-cultured cells as the starting material. A combination of these expertises may not be available in laboratories that either focuses solely on 3D cellular studies or concentrate in developing IF multiplexing exclusively for tissues, leading to the lack of published studies featuring IF multiplexing assays in 3D models.
To adopt 3D multiplexing as a diagnostic and prognostic tool, it is necessary to standardize and validate the staining and imaging processes. Manual fluorescent multiplexing protocols are not only laborious and time-consuming, they introduce errors and inconsistencies, and therefore difficult to validate. The use of automated IHC staining instruments and locked protocols in combination with spectrally unmixed multiplex images ensure less variability and faster processing. However, standardization is required at each step for a robust and reproducible system that is validation-worthy. While setting up a fluorescent IF multiplex system for cancer diagnostics, several protocol parameters of the tyramide-based automated staining system requires standardization. The choice of primary antibody (including characterization of its epitope) is of utmost importance as the starting material. Several steps in the automated protocols require individual standardization before combining all the parameters to generate a robust and reproducible multiplex IF assay. These include dilution of primary antibody (titration), primary and secondary antibody incubation time, choice of fluorophores, antibody-fluorophore pairing, the placement sequence of the antibody-fluorophore pairs in the staining protocol, and the sensitivity of antibody-fluorophore complexes to heat deactivation. In addition, the time of fluorophore deposition and the number of washes to remove primary-secondary antibody complexes to below-detection level requires optimization.
Our choice of biomarkers was driven by increased clinical needs to identify key proteins specific to different cancers and their subtypes. NCI-H2228 cells contain translocated ALK gene, and the ALK (D5F3) antibody detects total endogenous ALK, EML4-ALK fusion, and NPM-ALK fusion proteins in tissues that account for 3%-7% of NSCLC. Gain-of-function ALK gene rearrangements and EGFR gene mutations are single-gene predictive biomarkers for progression-free survival in treatment with tyrosine kinase inhibitors. Both NCI-H2228 (EGFR WT) and NCI-H1975 (EGFR mutations L858R and T790M) showed strong membranous to diffuse cytoplasmic staining with EGFR (5B7) antibody, which detects the EGFR intracellular domain. EGFR overexpression is found in 43%-89% of non–small cell lung adenocarcinoma, and protein expression using EGFR (5B7) antibody predicts better response to gefitinib in patients with NSCLC, including patients carrying EGFR mutations.42 TTF-1 is a predictive and prognostic biomarker whose expression is associated with advanced lung adenocarcinoma43 while CK7 is a cytoplasmic protein expressed in most non–metastatic primary lung adenocarcinoma.36
Defects in MMR proteins MSH2, MSH6, MLH1, and PMS2 contribute to MSI and increased tumor mutation burden. Loss of staining in any of the markers MSH2, MSH6, MLH1, or PMS2 signifies MSI and correlates with oncogenesis in various sites including colorectal, endometrium, ovary, pancreas, gastric, and brain.44 In CRC, germline mutation in one or more of the four MMR genes causes Lynch syndrome.45 While the KM12 cell line carries a mutation in MLH1, the other three genes, PMS2, MSH2, and MSH6 are wild-type. HT-29, a microsatellite stable cell line, carries the wild-type genes for all four MMR proteins. In advanced CRC, BRAF V600E mutation is associated with poor prognosis, as targeted therapies have limited success due to high heterogeneity within tumors.46 CK20 is a cytokeratin protein exclusively expressed in the cytoplasm and a marker of metastatic colorectal adenocarcinoma.35 While we focused on proteins affected by genetic alterations (ALK, EGFR, BRAF V600E, MSH2, and MSH6) in these studies, immune checkpoint biomarker(s) expressed on tumors (such as PD-L1) can be included with the repertoire of biomarkers. In addition, our multiplex IF system can accommodate more antibodies per panel and is not restricted to the 4-plex IF depicted in this study.
The 3D spheroid methodology described in this study can serve as a substitute for commonly used tissue microarray and xenografts for verification screening. The potential to visualize multiple biomarkers in their spatial context and the ability to study protein-protein interactions between them opens up a new paradigm in cancer preclinical studies. The workflow can be adapted to several applications of the 3D spheroid technology including cell viability and toxicity studies following drug treatment in cancer cell lines alone or with cocultured cancer-associated cells.47, 48 Other possibilities include human tumor-derived organoid cultures or patient-derived cultures49 and HTS50 of multiple biomarkers in a single well of a multiwell plate. In the case of patient tumor-derived 3D organoids, the expression status and spatial arrangement of multiple disease-relevant biomarkers can be studied in a preclinical nontumor microenvironment setting. An IHC-based screening strategy for late-stage NSCLC-derived 3D organoids would include IF multiplexing of key biomarker proteins such as ALK, EGFR, the MMR proteins MLH1, PMS2, MSH2, and MSH6, ROS1, BRAF V600E, NTRK, PD-L1, and TTF-1, to name a few, in various combinations, to study patient stratification approaches for different targeted therapies.
Image quantification of expressed proteins in a multiplex setting added an extra dimension to the understanding of biomarker expression dynamics in these model cell lines. Conventional visual-based, manually annotated, and counted image assessment does not have the ability to assess multiple biomarkers in large numbers of cells from several FOV. The automated scoring system enables the rapid processing of extensive datasets, thereby removing the limitations of time-consuming processes and imprecise estimates generated by manual semiquantitative and qualitative analyses. The automated analyses used in these studies enhance our potential to develop high-throughput 3D cell screening methods following drug treatment where rapid, accurate, and simultaneous computation of multiple expressed biomarkers is necessary to investigate treatment response and protein interaction networks.
ACKNOWLEDGMENTS
The authors would like to thank Scott Gill, Kevin Boone, Liliana Rounds, Krispy Saufkie, Erika Castiblanco, Pengfei Gu, Alyssa Jordan, Jennifer Cronkhite, Andre Zamorano, Maria Herrera, and Diana Bryant for help with reagents, slides, and chromogenic staining, Xiaoling Xia for advice, Karl Garsha for software support, and Laya Bhavaraju for manuscript management.
CONFLICT OF INTERESTS
Srabani Bhaumik, Jean Boyer, Chaitali Banerjee, Samantha Clark, Noemi Sebastiao, Elizabeth Vela, and Penny Towne are employees of Roche.
AUTHOR CONTRIBUTIONS
Conceptualization methodology, investigation, resources, formal analysis, and writing—original draft, review, and editing: SB. Conceptualization, methodology, investigation, resources, and writing—reviewing and editing: JB. Conceptualization, methodology, investigation, formal analysis, writing—original draft, reviewing, and editing: CB. Methodology and investigation: SC. Conceptualization, resources, and writing—review and editing: NS. Writing—review: EV. Funding acquisition, resources, and writing—review: PT.