Peptidyl inhibition of Spt4-Spt5: Protein-protein inhibitors for targeting the transcriptional pathway related to C9orf72 expansion repeats
Abstract
Spt4/Spt5 is an useful target as it is likely a transcription factor that has implications for long non-coding RNA repeats related to frontotemporal dementia (FTD) found in the C9orf72 disease pathology. Inhibitors for Spt4/Spt5 using peptides as a starting point for assays as a means for developing small molecules, which could likely lead to therapeutic development for inhibition for Spt4/Spt5 with CNS characteristics. To elucidate the specific steps of identification and modification of key interacting residues from Spt4/Spt5 complex with further effect prediction, a set of different computational methods was applied. Newly characterized, theoretically derived peptides docked on Spt4/Spt5 models, based on X-ray crystallography sources, allowed us to complete molecular dynamics simulations and docking studies for peptide libraries that give us high confident set of peptides for use to screen for Spt4/Spt5 inhibition. Several peptides with increased specificity to the Spt4/Spt5 interface were found and can be screened in cell-based assays and enzymatic assays for peptide screens that lead to small molecule campaigns. Spt4/Spt5 comprises an attractive target for neurological diseases, and applying these peptides into a screening campaign will promote the goal of therapeutic searches for FTD drug discovery.
1 INTRODUCTION
Transcription by RNA polymerase (RNAP) ranks first in gene expression, and this process is highly regulated at many steps, including promoter recognition, transcription activation, elongation, and termination. Elongation by RNA polymerase II (RNAPII) is a dynamic stage, engaging a set of proteins and forming really precise and coordinated machinery, where structural and functional components interact with each other.1, 2 After consideration of the network, it becomes evident that one of the most critical crossroads is the DRB sensitivity inducing factor (DSIF), which interacts with the positive transcription elongation factor b complex, RNA polymerase II, negative elongation factor complex, and many other partners.3 Human DSIF is composed of Spt4 (SUPT4H1) and Spt5 (SUPT5H). At first glance, the complex seems to be significant only for ensuring the stability of the elongation machinery on the chromatin and further activation or inhibition of transcription elongation process. However, Spt4/Spt5 also compensates changes in elongation activity by reducing the frequency of its pausing or arrest.4 Single Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences,5 while DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation.6 For example, in conditions of limited nucleotide number, the frequency of arrests and pauses increase dramatically, but in vitro, the complex Spt4/Spt5 promotes the transcription elongation process.7
In vitro studies on transcription mechanism,4 especially interaction with immunodeficiency virus type 1 Tat8 and modulation of HIV-1 replication by RNA interference directed against human transcription elongation factor Spt5,9 revealed that formation of Spt4/Spt5 complex begins just downstream of the transcription initiation site. Association of Spt4/Spt5 complex with other proteins shares its shape and functions between the “DNA exit tunnel” and the “RNA exit tunnelm” In this case, due to its external domains, Spt5 plays a crucial role in controlling transcription, as it is required to provide RNAPII transcription at a sufficient rate.
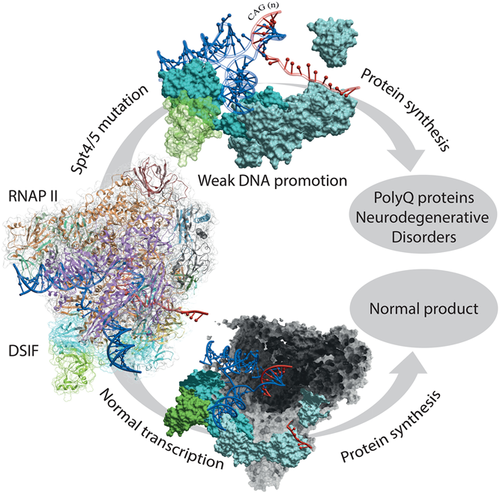
However, Spt4 should not be discounted as several mutations of Spt4 selectively decreased synthesis of polyQ by reducing dissociation of RNA polymerase II from the template. Such multiplied cytosine-adenine-guanine (CAG) repeats, encoding a long polyglutamine (polyQ) pattern, are known to become structurally unstable in proteins and prone to aggregate (Figure 1). Such polyQ disorders form a group of neurodegenerative diseases like Huntington's disease, Spinocerebellar Ataxia, and Machado-Joseph disease. Later studies support that prevention of the Spt4/Spt5 interactions affects the reduction of mutant polyQ protein accumulation10 positively. Therefore, inhibition of the protein-protein interaction (PPI) is an alternative and efficient way to treat many neurodegenerative diseases. Additionally, it now considered to be a favorite target for frontotemporal dementia resulting from C9orf72 expansion that results in long non–coding RNAs that get RAN translated into large plaque proteins consisting of GA, GR, or GP repeats.11, 12 Thus, finding inhibitors for Spt4/Spt5 is an essential drug target.
For now, there are different antibodies and peptide antigens, designed to inhibit Spt4/Spt5 association, which can bind to the individual subunits, and finally, arrest transcription to forestall mentioned neurodegenerative disorders. All these constructions, built on the mouse or rabbit systems, were designed, crudely, against three epitopes. The first group is focused on the full-length Spt4 of human origin, the second group is complementary to the N terminus of Spt5, and the third group covers those antibodies against CTR domain. All of them demonstrate a relatively effective inhibitory action. However, the production and following delivery of such large protein of about 120 kDa into the correct tissue or cell type is a slightly complicated and expensive task for the organism.13 There are several benefits of peptides over the whole protein, for example, lower probability of cross-reactivity and more flexible antigen selection, which requires only the protein sequence. Another advantage of peptide antigen design is a well-defined epitope from the beginning and fast turnaround time of the process in vitro. Thus, a peptide of 10 to 20 residues with a somehow increased inhibitory activity against any of the DSIF partners is an excellent biotechnological tool to counteract PolyQ proteins formation.
For the rational design of such peptides, we decided to realize the molecular recognition pattern of the PPI. To understand it, the interaction between Spt4 and Spt5 were in silico studied to determinate the hot spot regions of both peptides by molecular dynamics (MDs) simulation. Then a set of short peptides were proposed as promising PPI inhibitors and their potential activity was proved by a set of in silico modeling of Spt4 and Spt5 interaction with peptides proposed.
2 MATERIALS AND METHODS
Novel peptide design and computational modeling were based on the existing crystal of the Spt4/Spt5 complex (PDB code: 3H7H), demonstrating the PPI region of interest. Both proteins from the complex were chosen as targets source for peptide sequence extraction, studies on peptide binding, following simulations and analysis. As the original PPI interface shares similar secondary structure with a β-sheet folding, formed with a couple of single β strands from each protein, the idea was to enhance such interaction with point mutations and evaluate any changes with the analysis of MD simulations.
The algorithm of amino acid replacement was applied according to the reported statistical and experimental rules14 (Table 1). Thus, all modification within the rules had to increase the stability and affinity of the predicted peptide-protein interaction, namely β2 and β3 strands of Spt5 and Spt4, respectively. The fragment of 11 to 12 amino acids derived from the interface of each partner from of DSIF was used as a template for substitutions and comparison. All sequence alignments were carried out with Molsoft ICM tools15 and structural alignment and visualization with Pymol 1.5 software.16
| Source | Facing residues | |
|---|---|---|
| Non-H-bonded sites | H-bonded site | |
| Statistical data | C-C>>E-K>D-H>N-N>W-W>C-W≈D-G≈D-R>K-N≈N-S>H-P≈Q-R17 | C-C>E-K≈E-R>H-H≈Q-R≈D-N≈F-F≈C-H≈S-S≈D-K≈K-Q≈N-T17 |
| Experimental data | W-W>>W-F>W-Y>W-L>W-M>W-I>W-V>>Y-L>M-L>F-L>L-L>I-L≈V-L C-C18; Y-W19; I-W14 F-F20; Y-F | V-V>H-V≈V-H18 S-T, T-T19 N-T19 S-T14 |
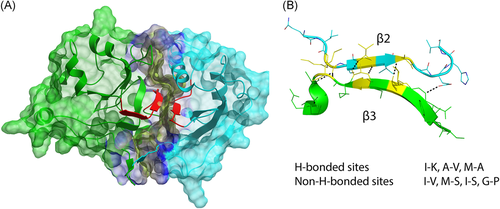
While developing novel peptides, we relied on the initial secondary structure of the β strand from peptide templates. That means that even without a β-turn secondary element a general representation of the required interaction between the protein and peptides should be represented with alternating H-bonding and non–H-bonding sites.21, 22 Poly-arginine tails were added to increase permeability due to the ability of the region to form pores within lipid bilayer.23 However, these tails could be cleaved out inside the cell using the introduction of the cleavage site in the sequence.24, 25 All peptides were slightly relaxed during 5 ns to reproduce the influence of the solvent and vary peptide conformations.
All the replacements and subsequent minimizations of the obtained peptide structures were performed with the Molsoft ICM tools. Redocking of novel peptides was provided with Global FFT dock and Legacy protocols to confirm the probability of the binding between peptide and the target protein. To increase the accuracy of the procedure, the known binding site of the opposite protein was determined as an epitope for docking.
All individual peptides and protein-peptide complexes were solvated with water solvent molecules (SPC216), specifying a solute-box distance of 1.0 nm. All steps of energy minimization and system equilibration MD simulations of 50 to 100 ns were performed using the Gromacs (ver 5.1.4) program and the GROMOS 53a6 force field19 at 310 K, using particle-mesh Ewald method for calculation of long-range interactions. Thermostat v-rescale for temperature coupling and Parrinello-Rahman barostat was used during the production run. Cutoff radii of 1 nm were applied for both the Coulomb (electrostatic) and the Lennard-Jones (Van der Waals) interactions. Calculation of binding free energy was performed with a more precise MM-PBSA method. The resulting structures generated after the production MD run were subjected to binding-free energy calculation. Each protein and the corresponding fragment of another one, from the Spt4/Spt5 complex, were processed using the MM-PBSA tool implemented in GROMACS.17
3 RESULTS
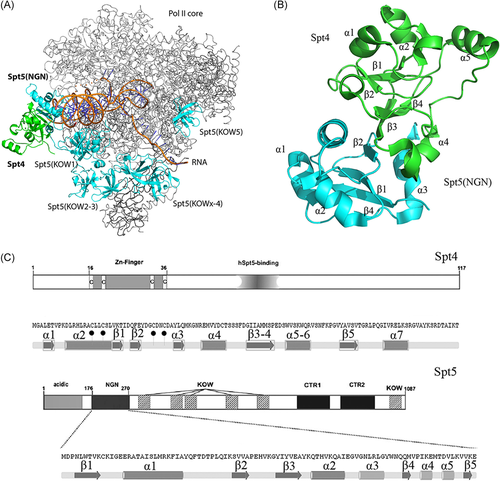
For the hot spots determination, the analysis of whole polymerase complex was performed to find the most promising Kyprides-Onzonis Woese (KOW) domain from Spt5, which were shown to provide protein-protein and protein-nucleic acid recognitions. Eukaryotic Spt5 is a multidomain protein composed of the conserved NusG N-terminal (NGN) domain, directly interacting with Spt4, and contains additional motifs: the N-terminal acidic region, four to five additional KOW motifs, and flexible linkers preceding a C-terminal repeat region (CTR). Initially, the idea was to design peptides against individual KOW domains or find the common interface to affect all of them at the moment, but their physical resemblance proved to be illusive therefore we decided to analyze the NGN role in the interaction.20, 26 In contrast to extended Spt5 subunit of DSIF, Spt4 partner is a small zinc-finger protein, which consists of an N-terminal 4-Cys Zn-finger, and exhibits α/β topology (Figure 2). It is important that mutation of any one of four cysteines from the zinc finger structure causes dangerous disorders.27-29 However, other, non-cysteine, point mutations disabling Spt4 zinc-finger domain functioning reduced PolyQ protein expression.18 Therefore, we focused our attention to a more deep investigation of the interaction pattern in the abovementioned domain.
For the identification of key amino acids residues for PPI, we analyzed the existing deposited structures of the DSIF complex. The central feature of the Spt4/Spt5 interface is a large β sheet formed by a combination of four antiparallel strands from each of Spt4 and Spt5, resembling a single large hydrophobic surface.30 Several α-helices are flanking the plane and contributing charged and polar interactions, which is necessary for Spt4 and Spt5 contacts. Mutations targeting such contacts between Spt4 and Spt5 disrupted Spt4/Spt5 interactions both in vitro and in vivo.31
Based on the analysis of the surfaces, the most structurally stable and, besides, short enough fragments represented with a “KSVVA” and “DGIIAM” sequences, from Spt5 and Spt4, are responsible for the PPI. These β strands are forming the β-sheet-like structure, where the core part consists of β2 and β3 strands of Spt5 and Spt4, which are flanked with α helices. Altogether these substructures impact the contact surface (Figure 3).
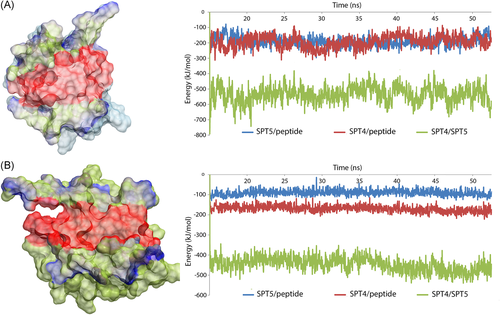
To prove the aforementioned compilation, a set of MD simulation was performed. For these, we constructed complexes containing each protein with a short corresponding peptide (fragment of the partner protein). First of all, the interaction surface is formed with disparate secondary structures, which are connected with disordered linkers and, probably, are stable because of the folding of the proteins. For example, one of the further designed peptides consisted of the β strand connected to a short α helix, it was extracted from the Spt4 structure and showed a significant destabilization of the whole fragment during MD in a water solution. Next step assumed a long-range MD simulation of the complex during 50 ns with subsequent energy analysis and comparison of the impact of different secondary structure elements engaged in the PPI. These data showed that 10 residues of each protein, forming β2 and β3 strands, contribute about the quarter of all electrostatic and van der Waals (VdW) interactions of the surface. As a result, we concluded that the areas around both β strands involved in the contact, shown with red in Figure 4, are the most important and promising binding sites for newly designed peptides along with the fact that these sequences should become a source for modifications.
For the generation of potent peptide-based PPI inhibitors, we used two “KSVVA” and “DGIIAM” sequences as a starting point for pseudocombinatorial modification. The amino acid replacement was made in accordance with the data from Pantoja-Uceda et al14 leading to more than 20 different sequences of each fragment bearing polyarginine tails. The sequence length up to 16 residues was considered to be enough for interaction and potential synthesis. As the core of each interface is a β strand, a set of statistical and experimental rules were implied to modify the source sequence to increase the regularity of the peptide along the interaction axis. A variety of peptide length was also taken into account because the value can affect the size of the disordered area, resulting in the reduction of interaction energy strength. Both length and the sequence are responsible for inner strain, flexibility, and finally affinity of the peptide. It is especially notable in the case of a shallow cavity or extended structure of the site, as it is in the Spt4/Spt5 complex.
In addition to the β-strand part, another element of such secondary structure is proline/glycine-rich β turn or hairpin, which is also common for PPI inhibitors. It is important to focus on D-amino acids, for example, proline, as they could change the entire conformation of a single strand bending in another way. At the same time such exchanges have a positive effect on β-sheet structure stability.
A Global FFT protein-protein docking approach was used to generate protein-peptide complexes and simulate the validity of its configuration. The best way to estimate the binding probability is docking of the peptide from the short free MD simulation in the water box with counter ions to neutralize a non-zero charge of the protein and cross the energy barrier, caused with amino acid replacement. These MD simulations last for 5 ns to relax the structure, but not to study the protein behavior in a more detailed manner. Based on the clustering of MD trajectories, a set of 1 to 3 conformations were extracted and then used in a protein-protein docking for the complexes and likewise for other purposes.
A complex formation is considered to be possible if the peptide's structure does not undergo drastic transitions (conformational changes) during the MD simulation and then protein-protein docking shows a binding mode that is similar to the reference protein's fragment position. However, the dissociation probability can only be estimated with the resulting MD analysis.
This is currently considered the most correct validation protocol to follow for a rigorous peptide-protein and protein-protein system. To evaluate if the novel complex is stable and demonstrates increased "tight contacts" and appropriate energy values, a series of MD simulations were run for 100 ns each and then analyzed.
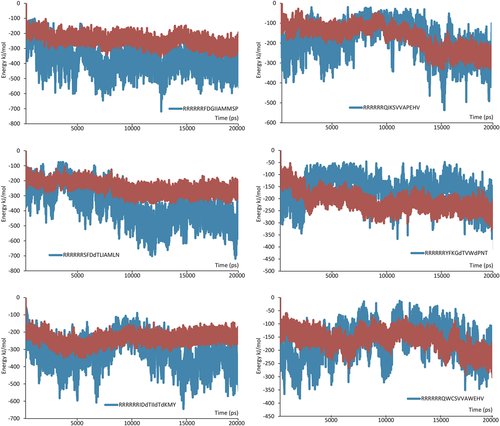
To demonstrate the probability of a tight interaction, which prevents the natural complex formation via competitive binding, a series of MD simulations were carried out for each case and then analyzed. The best combination of calculated electrostatics and VdW components assisted in forming the trend in the series of peptides. Several examples of interaction energy analysis are shown, in Figure 5, where each belongs to the most stable protein-peptide complex. However, as we mentioned before, several complexes were declined, despite sufficient positive data because the extracted peptide ligand must also be able to keep a stable conformation during the simulation in the water box. If the peptide fails to maintain stability during the simulation, then it too must be discarded. Based on the obtained results, only six short peptides were found to be the most promising and, thus, were chosen for further investigation.
Selected peptides were analyzed to count hydrogen bond number changes during the MD simulations between two protein units. The data added to our visual inspections of the PPI, reflects the overall picture of the associated peptides affinity and stability (Figure 6).
β sheet regularity of the designed peptides in complex with a partner protein was estimated based on the analysis of dihedrals during MD with DSSP-based32 secondary structure assignment, RMSD of the peptide structure and hydrogen bonds, in other words, a consistency of HB and non–HB sites of pseudo-β-hairpin. Binding energy between each pair of peptides and proteins was calculated with a PBSA method (Table 2).17 Due to a high degree of flexibility of the polyR tail, which influences the result of the calculation, and the probability of insertion of a cleavage site, the only target peptides' sequences were processed.
| Sequence | Target | H bonds | β-Sheet state occupancy, % | Binding energy, kJ/mol |
|---|---|---|---|---|
| (R)5IDdTIIdTdKMY | Spt4 | 5.020 | 65 | −116.758 |
| (R)5SFDdTLIAMLN | Spt4 | 11.150 | 94 | −148.381 |
| (R)5FDGIIAMMSP | Spt4 | 10.007 | 100 | −132.790 |
| (R)5QWCSVVAWEHV | Spt5 | 5.106 | 39 | −68.758 |
| (R)5YFKGdTVWdPNT | Spt5 | 5.895 | 66 | −147.022 |
| (R)5QIKSVVAPEHV | Spt5 | 5.196 | 78 | −118.378 |
- Note: Each red-colored area, which surrounded by a dark blue halo, corresponds to the entire binding region on the surface.
4 CONCLUSION
A set of modified peptides was designed to be more complementary against each protein from the DSIF complex. The most crucial residues, which make a larger contribution in the PPI of Spt4/Spt5, were identified, and their role was proved with a MDs simulation. Using the initial single-point mutation method, we let the peptide grow via extended replacement, which significantly increased the predicted binding effect. A cross-analysis determination of each subunit of DSIF allowed the generation of 20 short candidates of peptides. Predicted binding modes were determined through molecular docking and MD simulations. Both, a probable degree of stability and the estimated potency (level of dissociation) were evaluated using Gromacs' built-in analysis tools and then represented with a graphic of interaction energy fluctuations, which fully reveals the difference for each group of designed peptides.
Finally, several of tested peptide samples showed even better results than the original sequence derived from the protein complex. The quality of selected peptides was also analyzed with binding energy analysis and a subsequent DSSP analysis. Based on a set of obtained parameters, the most promising peptides can be used for in vitro study moving closer to novel peptide-based means of blocking the aberrant translation of the deadly expansion repeats found in C9orf72.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.