4-Hydroxy-2-nonenal attenuates 8-oxoguanine DNA glycosylase 1 activity
Guodong Pan and Mandar Deshpande contributed equally to this study.
Abstract
Elevated cellular oxidative stress and oxidative DNA damage are key contributors to impaired cardiac function in diabetes. During chronic inflammation, reactive oxygen species (ROS)-induced lipid peroxidation results in the formation of reactive aldehydes, foremost of which is 4-hydroxy-2-nonenal (4HNE). 4HNE forms covalent adducts with proteins, negatively impacting cellular protein function. During conditions of elevated oxidative stress, oxidative DNA damage such as modification by 8-hydroxydeoxyguanosine (8OHdG) is repaired by 8-oxoguanine glycosylase-1 (OGG-1). Based on these facts, we hypothesized that 4HNE forms adducts with OGG-1 inhibiting its activity, and thus, increases the levels of 8OHG in diabetic heart tissues. To test our hypothesis, we evaluated OGG-1 activity, 8OHG and 4HNE in the hearts of leptin receptor deficient db/db mice, a type-2 diabetic model. We also treated the recombinant OGG-1 with 4HNE to measure direct adduction. We found decreased OGG-1 activity (P > .05), increased 8OHG (P > .05) and increased 4HNE adducts (P > .05) along with low aldehyde dehydrogenase-2 activity (P > .05). The increased colocalization of OGG-1 and 4HNE in cardiomyocytes suggest 4HNE adduction on OGG-1. Furthermore, colocalization of 8OHG and OGG-1 with mitochondrial markers TOM 20 and aconitase, respectively, indicated significant levels of oxidatively-induced mtDNA damage and implicated a role for mitochondrial OGG-1 function. In vitro exposure of recombinant OGG-1 (rOGG-1) with increasing concentrations of 4HNE resulted in a concentration-dependent decrease in OGG-1 activity. Mass spectral analysis of trypsin digests of 4HNE-treated rOGG-1 identified 4HNE adducts on C28, C75, C163, H179, H237, C241, K249, H270, and H282. In silico molecular modeling of 4HNE-K249 OGG-1 and 4HNE-H270 OGG-1 mechanistically supported 4HNE-mediated enzymatic inhibition of OGG-1. In conclusion, these data support the hypothesis that inhibition of OGG-1 by direct modification by 4HNE contributes to decreased OGG-1 activity and increased 8OHG-modified DNA that are present in the diabetic heart.
1 INTRODUCTION
The augmented reactive oxygen species (ROS) generation in pathophysiological conditions cause oxidative stress.1 Oxidative stress leads to damage, dysfunction, and disturbances in cellular structures starting from the outer plasma membrane to inner nuclear DNA2 through a variety of mechanisms. One of the mechanisms is through reactive aldehydes like 4-hydroxy-2-nonenal (4HNE), which are generated upon lipid peroxidation. Lipid peroxides such as 4HNE can form covalent adducts with cellular proteins on cysteine, lysine, and histidine residues, thereby impairing function.3, 4 If cellular antioxidant defenses are inadequate, these modifications can lead to cellular stress and dysfunction. In general, oxidized proteins and RNA are destroyed by cellular machinery. However, oxidative DNA damage is repaired as it is essential for genomic informational integrity.5 Oxidative DNA damage can occur on different components of DNA molecules including bases, sugar fragments, abasic/apurinic/apyrimidinic (AP) sites, and single/double-strand breaks.6 Among nucleotides, guanine is more susceptible to modification by reactive oxidative intermediates.7 Oxidative modification of guanine results in the formation of 8-hydroxyguanine (8OHG), 8-hydroxydeoxyguanosine (8OHdG), and 8-oxo-7,8-dihydro-2-deoxyguanosine (8-oxodG). Importantly, these oxidized DNA molecules are established markers of ROS-induced DNA damage.8
8-oxoG is one of the most commonly formed oxidative lesions in cells.9 It mispairs with A during replication, thus making it a highly mutagenic lesion.10 Base excision repair (BER) is a primary DNA repair mechanisms.11 Recently, there has been a lot of renewed interest developed in DNA repair mechanisms with this topic receiving the 2015 Nobel Prize for Chemistry. 8-oxoguanine glycosylases (OGGs) is a group of eight DNA repair glycosylases. OGG-1 identifies and excises 8OHG and 8-oxoG and imidazole ring-fragmented guanines (FapyG) lesions.12 It is a bifunctional enzyme: in addition to glycosylase activity, it has lyase activity.13 First OGG-1 flips the damaged base out of the DNA double helix and cleaves the N-glycosidic bond of the damaged base (thus called glycosylases), leaving an AP site.14 Next, OGG-1 executes its lyase activity, that is, cutting the phosphodiester bond of DNA, creating a single-strand break.15
An increase in steady-state levels of 8-oxodG in tissues of OGG-1 knockout mice compared with its wild-type counterparts implies the importance of OGG-1 activity in keeping DNA damage in check.16 The decrease in OGG-1 activity has been implicated in variety of diseases including cancers, neuro degenerative diseases, and metabolic conditions as oxidative stress is prevalent in those diseases.17 Diabetes-mediated 8-oxodG levels were increased in the kidney tissue of type-1 diabetic rats.18 Overexpression of OGG-1 prevented fibrosis and ameliorated the type-1 diabetic heart.19 Using a high-fat diet model, Sampath et al showed increased lipid accumulation in the liver of OGG-1 knockout mice compared with their wild-type counterparts.20 In type-1 and type-2 diabetic models, we found increased 4-hydroxy-2-nonenal (4HNE) protein adducts in diabetic tissue.21, 22 Previous studies support aberrant posttranslational modification of OGG-1 contributes to decreased OGG-1 activity: in type 1 diabetic mice, O-GlcNAcylation of OGG-1 was increased corresponding to decreased OGG-1 activity.23 Therefore, taken altogether, we hypothesize that 4HNE decreases OGG-1 activity by forming adducts with OGG-1 on critical sites in diabetic tissue. We tested our hypothesis using recombinant OGG-1 in vitro and in the heart tissues of db/db mice, a type-2 diabetic model.
2 MATERIALS AND METHODS
2.1 Experimental animals
Leptin receptor mutant mice, db/db (n = 12) and their controls, db/dm with C57BL/6 background (n = 12) were purchased from Jackson Laboratory (Bar Harbor, ME). After 6 to 9 months, the animals were killed, and the heart tissue was isolated. They were appropriately stored for biochemical and histopathological studies. The animal protocols were approved by the Henry Ford Health System Institutional Animal Care and Use Committee. They adhere to the guiding principles of the care and use of experimental animals in accordance with the NIH guidelines. Henry Ford Hospital operates an AAALAC certified animal facility.
2.2 Immunostaining of 8OHG
Formalin-fixed, paraffin-embedded cardiac tissue sections were used for immunostaining. After deparaffinization and hydration, the slides were washed in Tris-buffered saline (TBS; 10 mmol/L Tris-HCl, 0.85% NaCl, pH 7.5) containing 0.1% bovine serum albumin (BSA). Antigen retrieval was performed using the pressure cooker method at 110°C for 10 minutes. After overnight incubation with mouse 8OHG antibody (1:200; Abcam, Cambridge, MA) at 4°C, the slides were washed in TBS. Then we incubated with goat anti-mouse IgG Alexa Fluor 488 secondary antibody (Thermo Fisher Scientific, Waltham, MA) for 1 hour in room temperature. After mounting, the stained cardiac sections were visualized with the help of a microscope using an Olympus IX81 (Olympus America, Center Valley, PA). The images were captured with a digital camera (DP72) and DPC controller software (Olympus America) set at 488 nm excitation at ×40 magnification. We analyzed at least 10 high power images per cardiac section and at least 3 samples per group for quantification.
2.3 Immuno colocalization of 4HNE with OGG-1 and 8OHG
Following the standard deparaffinization protocol, colocalization of 4HNE with OGG1 and 4HNE with 8OHG was performed by using double immunostaining with 4HNE-Cys/His/Lys (Millipore Sigma) rabbit antibody (1:200) and OGG-1 mouse monoclonal IgG antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA). Then, we used the secondary antibodies anti-rabbit IgG Alexa Fluor 568 for red and anti-mouse IgG Alexa Fluor 488 for green (Thermo Fisher Scientific), respectively. Next, for the colocalization of 4HNE with 8OHG, 4HNE-Cys/His/Lys rabbit antibody (1:100; Millipore Sigma) and anti-mouse 8OHG antibody (1:100; Abcam) with the corresponding secondary antibodies, goat anti-rabbit IgG Alexa Fluor 488 for green and donkey anti-mouse IgG Alexa Fluor 568 for red were used. Histologic micrograph images were captured using an Olympus IX81 (Olympus America, Center Valley, PA). The images were captured with a digital camera (DP72) and DPC controller software (Olympus America) set at ×40 magnification. The details of fluorescent single band filters are as follows:
Blue (DAPI) excitation wavelength (Ex): 360 to 370 nm and emission wavelength (Em): 420 to 460 nm; Green (Alexa Flour 488) Ex: 460 to 500 nm and Em: 510 to 560 nm; Red (Alexa Flour 488) Ex: 533 to 587 nm and Em: 607 to 682 nm. These are the same for all florescence imaging done.
2.4 OGG-1 expression and purification
Human OGG-1 was expressed and purified from His-tagged constructs, as previously described and was the gift of Dr. R. Stephen Lloyd, Oregon Health and Science University.24 Purified preparations of OGG1 (>95% pure) were stored at −80°C in 20 mM Tris-HCl (pH 7.0), 100 mM KCl, and 10 mM β-mercaptoethanol, and 50% glycerol.
2.5 4HNE treatment and OGG-1 activity assay in vitro
The fluorescence-based assay was performed in black 96-well plates with a final volume of 100 μL/well by following the method described by Donley et al24 Assay buffer (20 mM Tris-HCl, 100 mM KCl, 0.1% BSA, 0.01% Tween-20, pH 7.5) was added to each well (made up to 100 μL), followed by addition of 1 μL of OGG-1 (prediluted in assay buffer + 0.15% Tween) or 200 μg of cell protein, or 4HNE and dimethyl sulfoxide in some wells, followed by 1 μL of 2.5 μM double-stranded DNA substrate (prediluted in assay buffer + 0.15% Tween). The DNA substrate contained a 5′ FAM with an 8-oxoG in at nucleotide 6, annealed to a complementary strand containing a 3′ HQ-1 (custom synthesized by Integrated DNA Technologies (IDT, Coralville, Iowa). The 4HNE (100 μM) treated wells were first incubated with OGG-1 and then with 4HNE for 1 hour at room temperature, before the addition of DNA substrate. The final concentration of each component in the reaction was 100 μM 4HNE, 50 nM enzyme, and 25 nM substrate. After the addition of the DNA substrate, the plates were incubated at room temperature for 10 minutes, and then FAM fluorescence was measured in a Synergy H1 plate reader (Ex, 494; Em, 520) for 1 hour, with readings taken at every 20 minutes interval. Total OGG-1 activity per well was calculated by background-subtraction (buffer and buffer + substrate DNA) of fluorescence values from each well.
We also measured OGG-1 activity in the cardiac tissue homogenate from control and diabetic mice using the same assay.
2.6 Western immunoblotting
The Western blot analysis was performed as described earlier.22, 25 In brief, protein samples from heart tissue homogenate were separated on sodium dodecyl sulfate polyacrylamide gels by electrophoresis. The proteins were then transferred to immobilon-P membranes (Millipore, Billerica, MA). 4HNE-protein adduct levels were determined using antibodies of anti-4HNE-Cys/His/Lys rabbit antibody (1:1000; Millipore Sigma, Burlington, MA) and porin mouse monoclonal antibody (1:2000; Abcam) was used as a housekeeping marker, for comparison. The bound primary antibodies were added to horseradish peroxidase (HRP)-coupled respective secondary antibodies, and then visualized by chemiluminescence detection reagents.
2.7 Immuno colocalization of 8OHG and OGG-1 with mitochondrial markers
Cardiac paraffin sections were deparaffinized, as mentioned above. Then, the coimmuno localization of 8OHG and OGG-1 with mitochondrial markers TOM20 and aconitase, respectively, was performed. First, the double immunostaining with primary antibodies of 8OHG; anti-mouse 8OHG antibody (1:100; Abcam) and Tom 20; anti-rabbit TOM 20 antibody (1:100; Santa Cruz) was used along with the respective secondary antibodies, anti-mouse IgG Alexa Fluor 488 for green and anti-rabbit IgG Alexa Fluor 568 for red (Thermo Fischer Scientific). Similarly, for the double staining of OGG-1 and aconitase, the primary antibodies: anti-rabbit OGG-1 antibody (Novus Biologicals, LLC, Centennial, CO) and anti-mouse aconitase antibody (Abcam) along with the respective secondary antibodies, anti-rabbit IgG Alexa Fluor 488 for green and anti-mouse IgG Alexa Fluor 568 for red (Thermo Fischer Scientific) were used. See above for microscopic and imaging details.
2.8 Mass spectrometry analysis of recombinant OGG-1 incubated with 4HNE
Recombinant OGG-1 protein was treated with 0, 10, or 100 µM 4HNE and 10 μg aliquots of each were buffered with 50 mM ammonium bicarbonate (AMBIC). 4HNE adducts were stabilized by reaction with 5 mM NaBH4 in equimolar NaOH. Following the NaBH4 reaction, cysteines were reduced with 5 mM dithiothreitol (DTT) then alkylated with 15 mM iodoacetamide (IAA). Excess IAA was quenched with a second addition of 5 mM DTT. Samples were digested at 37°C overnight using sequencing-grade trypsin (Promega, Madison, WI). Following digestion, all samples were speedvac to dryness and solubilized in 0.1% FA in preparation for LC-MS/MS. All analyses were made using an Acclaim PepMap RSLC, 75 μm × 25 cm column with LC-MS/MS performed on a Thermo Fusion Orbitrap mass spectrometer. 35-minute LC gradients were used, MS1 scans were collected at 120 K resolution and the most abundant species were selected for MS2 analysis by CID in the ion trap.
2.9 Protein and modified site identification
Mass spectrometer raw files were searched using a PEAKS Studio 8.5 build 20180507 against the Uniprot/TrEMBL human database (20 145 entries) using 10 ppm precursor mass tolerance and 0.6 Da product mass tolerance against a human database. PEAKS PTM was used to identify modified peptides using a panel of 313 variable modifications including 4HNE on C, H, and K (156.1150 Da addition). Carbamidomethylated C (57.0215 Da addition) was also set as a variable modification. PTM localization confidence was determined by PEAKS PTM. Quantification was done using a chromatographic peak area for the precursor. Data analysis was done in R v3.4.3 and Excel.
2.10 Molecular modeling
All simulations were performed using Discovery Studio version 2.5.5 (Accelrys Inc., San Diego, CA). The crystallographic coordinates of the 3.1 Å human 8-oxoguanine glycosylase distally cross-linked to guanine crystal structure (Protein Data Bank entry 3ih726) were obtained from the Protein Data Bank (http://wwww.rcsb.org). The 4HNE modification of the K249 and H270 residues of OGG-126, 27 was completed as follows: The resulting structure was then subjected to energy minimization utilizing a conjugate gradient minimization protocol (10 000 iterations) with a CHARMM Force Field and the Generalized Born implicit solvent model with simple switching.28 Schiff base adducts of 4HNE were built onto K249 and H270 as identified by MS/MS analysis and both the native and adducted models subjected to a further round of minimization as described above.
2.11 Statistical analysis
Data are presented as mean ± standard error of the mean (SEM). The Student t test was applied to compare the two groups in the studies.
3 RESULTS
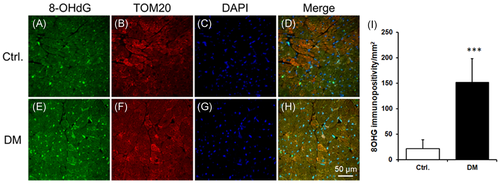
3.1 Diabetes increases 8OHG levels in the mitochondria of the heart tissue
8OHG levels were increased in the db/db mouse heart compared with db/dm mouse hearts (Figure 1A,C,D,G,E,H,I). Moreover, 8OHG was colocalized with the mitochondria as shown by TOM20, a mitochondrial protein (Figure 1B,C,D,F,G,H).
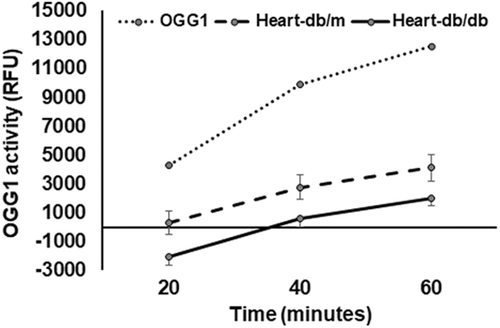
3.2 Diabetes decreases OGG-1 activity in heart tissue
OGG-1 activity was reduced in the db/db diabetic hearts compared that of db/dm hearts (Figure 2).
A significant amount of OGG-1 was colocalized with mitochondrial marker protein aconitase which is suggesting mtOGG-1.
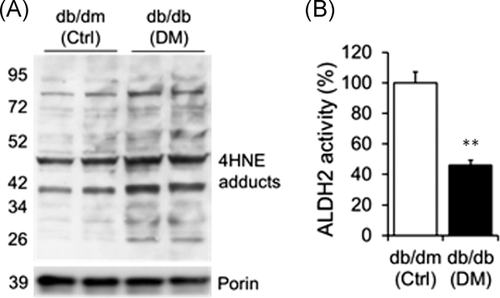
3.3 4HNE adducts are increased along with decrease in aldehyde dehydrogenase-2 activity in diabetic heart
Increased 4HNE adducts (Figure 3A) and decreased aldehyde dehydrogenase-2 (ALDH2) activity (Figure 3B) in db/db hearts were found relative to db/dm control hearts.
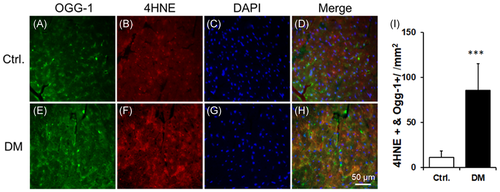
3.4 4HNE adducts are increased in OGG-1 in diabetic heart tissues
Increases in 4HNE-protein adducts on OGG-1 protein were found in the db/db hearts compared with the db/dm hearts (Figure 4A-I).
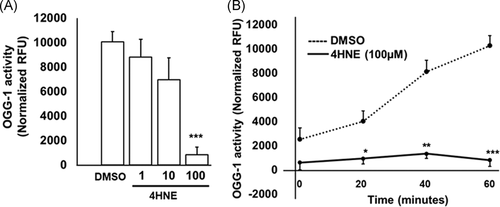
3.5 Exogenous 4HNE attenuates recombinant OGG-1 activity
Recombinant OGG-1 activity was reduced with 4HNE (1, 10, and 100 μM) in a dose-dependent manner (Figure 5A). Recombinant OGG-1 activity was lower in 20, 40 and 60 minutes with 100 μM 4HNE compared with vehicle treatment (Figure 5B).
3.6 4HNE adduct formation in OGG1 by mass spectroscopy
Treatment of recombinant OGG1 with 4HNE resulted in the formation of HNE adducts (Figure 6A). No 4HNE adducts were detected in untreated samples indicating our analysis is specific. Peptide abundances for HNE modified peptides in treated samples were determined using label-free quantification (Figure 6B).

3.7 Identification of 4HNE adducted residues in OGG-1
We identified C28, C75, C163, H179, H237, C241, K249, H270 and H282 in the OGG-1 protein structure (Figure 7).

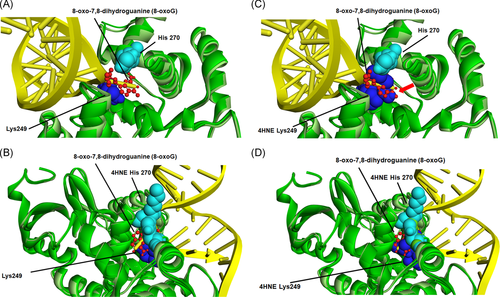
3.8 Molecular structural analysis of 4HNE adduction on OGG-1
The relative location of each adducted amino acid identified and its special relation to the DNA binding sites was first examined using a known crystal structure of OGG-1 (Figure S3).26 From the special comparison as well as that fact that both K249 and H270 have previously been demonstrated to be important in OGG-1 activity,27, 29 computational-based minimization simulations were performed focusing on the aforementioned residues using the crystal structure of OGG-1 bound to 8-oxo-7,8-dihydroguanine (8-oxoG) modified DNA (3ih726). Using in silico molecular modeling, 4HNE was adducted to K249 and H270 on human OGG-1. From the prediction of amino acid pKa values, H270 is protonated resulting in increased susceptibility to adduction. These results demonstrate that the charged nitrogen is the more likely target and addition of the 4HNE would abrogate the charge on the Histidine residue, impacting the interaction with the DNA backbone (Figure 8A,B). Examining adduction of K249, the addition of 4HNE alters both the positioning of the base of the DNA in the catalytic pocket, and abrogates the strong pi-cation interactions that K249 has with the base itself (Figure 8A,C—red arrow). This would directly impact enzymatic activity correlating with data demonstrating a 4HNE concentration-dependent inhibition of OGG-1 activity. Surprisingly, adduction of K249 did not impact the formation of H-bonds between K249 and Cys253 (Figure S4). This was also not impacted by the theoretical simultaneous double adduction of both K249 and H270 (Figure 8D). This would directly impact enzymatic activity correlating with data demonstrating a 4HNE concentration-dependent inhibition of OGG-1 activity.
4 DISCUSSION
In this study, we found increased 4HNE adduction on OGG-1 and reduction in its activity in type-2 diabetic heart tissue along with enhanced oxidatively-induced mitochondrial DNA damage, that is, increased 8OHG levels. Moreover, exogenous 4HNE attenuates OGG-1 activity by forming adducts on critical amino acids in the OGG-1.
Diabetes and its complications are a major healthcare issue around the globe and ~65% of diabetic people die of heart diseases. As oxidative stress has been identified as an important contributing factor for the diabetic heart diseases, we plan to focus on 4HNE-mediated molecular mechanism in OGG-1 impairment and resultant 8OHG levels.
An increase in DNA oxidative damage in the liver of streptozotocin (STZ)-induced type-1 diabetic rat was reported earlier.30 In another report using the same STZ model, it was shown that the OGG-1 level was reduced in the kidney cortex.18 However, our study is first to report increased oxidative mitochondrial DNA damage in the heart of db/db mice, a model of type-2 diabetic mice. In a previous study, by feeding a high-fat diet to OGG-1 knockout mice, Sampath et al20 found that they develop glucose intolerance, lipid accumulation in the liver and obesity compared with their wild-type counterparts. This study implicates the role of OGG-1 in the development of type-2 diabetes. A genetic polymorphism on OGG-1, that is S326C OGG-1 is associated with type 2 diabetes in humans.31 Our objective was to establish the association between mtOGG-1 activity and 8OHG levels in the type-2 diabetic heart.
We found that the 8OHG, an oxidized DNA molecule and TOM20, a mitochondrial marker protein, colocalized in diabetic heart tissue, implicating it in mtDNA damage. This finding is in accordance with another report in which they found a significant increase in 8-oxodG in the mitochondrial fractions of the kidney cortex of STZ-mediated type-1 diabetic rats, but not the nuclear fractions.18 We also found that OGG-1 was colocalized with aconitase, another mitochondrial marker, again implicating we have mtOGG-1. Although we found OGG-1 activity was significantly decreased in diabetic hearts compared with control hearts, we did not observe any decrease in mtOGG-1 levels in diabetic hearts compared with nondiabetic control hearts. This made us look for a novel mechanism for OGG-1 impairment in type 2 diabetic heart.
The attack by free radicals on biological membranes is called lipid peroxidation, which produces reactive aldehydes like 4HNE. 4HNE forms an adduct with macromolecules primarily with proteins and thereby affects their function. In this study, we found that an increase in 4HNE-protein adducts in diabetic heart tissue by both Western blot analysis and immunostaining. Additionally, we also found a decrease in the activity of ALDH2, a mitochondrial enzyme that detoxifies 4HNE. This may be another reason why these 4HNE adduct levels are increased in the diabetic heart. We also found this 4HNE adducts are colocalized with OGG-1 (Figure 3) and 8OHG (Figure S2), which implicates the 4HNE adduct formation in OGG-1, which perhaps increased 8OHG due to reduced OGG-1 activity due to 4HNE adduction on critical sites. In an earlier report, it has been shown that in the vulnerable regions of the brains of patients with Alzheimer's disease, OGG-1 activity is reduced due to 4HNE adduction.32 In this study, it was both mitochondrial and nuclear OGG-1. However, it is unknown at which sites 4HNE forms adducts with OGG-1. To determine this, we performed a few in vitro studies with recombinant OGG-1 activity. Treatment with 4HNE (1, 10, and 100 µM) attenuated OGG-1 activity in a dose-dependent manner in vitro. Montine et al have incubated the control brain lysates that had the highest OGG-1 activity with 5 µM 4HNE for 1 hour and found that OGG-1 activity was decreased ~50% implicating 4HNE can form adducts with OGG-1 readily and inhibits its activity significantly.
As we found 10 and 100 µM decreased OGG-1 activity significantly, we then incubated OGG-1 with 10 to 100 µM 4HNE and performed mass spectroscopy. As expected, we found 4HNE modifications in specific peptides with 10 and 100 µM 4HNE compared with control. We identified four such peptides. Next, we identified specific amino acid residues in those peptides which had 4HNE adductions. They included the amino acids C28, C75, C163, H179, H237, C241, K249, H270, and H282. Among these residues, K249 is known as a catalytic site for OGG-1. It is important to understand the biochemistry of these sites in terms of how base excision repair by OGG-1 is executed by recognizing and discriminating against the DNA lesions. There was a previous study published on mutations with some critical residues of OGG-1 and their importance on accessing and repairing the oxidized DNA such as 8-oxodG.29 In that study, they have also mutated H270 and K249. The inference from these mutations is: (1) H270A mutation diminishes the affinity of hOGG-1 for tetrahydrofuranyl-containing DNA by less than two-fold. (2) K249Q, a catalytic nucleophile did not interact with C253. This interaction is essential for the specific recognition of oxoG.29 Based on this biochemical structural study, we presume that the decrease in OGG-1 activity in diabetic heart tissue was due to 4HNE adductions on these critical sites.
Next, we performed in silico analysis to build molecular modeling simulations of the 4HNE adductions on those amino acids of OGG-1 along with its substrate to understand how 4HNE adductions can affect the function of OGG-1 at the biochemical level. Molecular modelling revealed two possible mechanisms of 4HNE-mediated attenuation of OGG-1 activity. First, modification of H270 would impact the ability of OGG-1 to associate with the DNA backbone. Second, adduction of K249 would impact OGG-1/8-oxoG substrate interactions and therefore catalysis.
In summary, in this report we have, for the first time, found mtOGG-1 activity is reduced, which increased 8OHG in the mitochondria of cardiac tissue procured from diabetic db/db mice. Concurrently, from in vitro studies using recombinant protein, we have identified 4HNE adduction sites on the OGG-1 and determined plausible mechanisms of 4HNE-mediate OGG-1 inhibition. Thus, a future goal is determining whether protecting those specific sites from 4HNE adduction can decrease mtOGG-1 inactivation. As the mitochondrial ALDH2 detoxifies 4HNE, activation of ALDH2 may be an effective strategy to improve mtOGG-1 activity in the diabetic heart. Finally, in conclusion, we propose that 4HNE attenuates mtOOG1 activity by forming adducts with critical amino acids, which can be decreased by enhancing 4HNE detoxification.
ACKNOWLEDGMENTS
We thank Oskar Linde and Stephan Lloyd from Oregon Institute of the Occupational Health Sciences, Oregon Health & Science University Portland, Oregon for the gift of recombinant hOGG-1 protein. This study was partially supported by the National Heart, Lung, and Blood Institute 1R01HL139877-01A1 (SSP), the National Heart, Lung, and Blood Institute 1 R56 HL131891-01A1 (SSP) and an internal grant from the Henry Ford Health System: A10249 (SSP). the National Institute of Alcohol Abuse and Alcoholism R37AA009300-22 (CTS). The Wayne State University Proteomics Core Facility is supported by NIH grants P30 ES020957, P30 CA 022453, and S10 OD010700.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
GP: planned and conducted the animal experiments, performed microscopy and some part of the biochemistry and made figures. MD: planned and conducted the biochemical experiments, involved in figure making and contributed to manuscript writing and editing. HP: immunostaining. PMS: planned and conducted mass spec studies. NJC: analyzed the mass spec data, made figures and contributed to manuscript writing. CTS: planned and conducted molecular modeling studies, made figures and contributed to manuscript writing and editing. DSB: planned and conducted molecular modeling studies and made figures. SSP: conceived the idea, planned the study, obtained the funding, supervised the experimentation and data analysis, wrote the manuscript and editing.