Pancreatic stellate cell-potentiated insulin secretion from Min6 cells is independent of interleukin 6-mediated pathway
Abstract
Pancreatic stellate cells (PSCs) secrete various factors, which can influence the β-cell function. The identification of stellate cell infiltration into the islets in pancreatic diseases suggests possible existence of cross-talk between these cells. To elucidate the influence of PSCs on β-cell function, mouse PSCs were cocultured with Min6 cells using the Transwell inserts. Glucose-stimulated insulin secretion from Min6 cells in response to PSCs was quantified by enzyme-linked immunosorbent assay and insulin gene expression was measured by quantitative polymerase chain reaction. Upon cytometric identification of IL6 in PSC culture supernatants, Min6 cells were cultured with IL6 to assess its influence on the insulin secretion and gene expression. PLC-IP3 pathway inhibitors were added in the cocultures, to determine the influence of PSC-secreted IL6 on Glucose-stimulated insulin secretion from Min6 cells. Increased insulin secretion with a concomitant decrease in total insulin content was noticed in PSC-cocultured Min6 cells. Although increased GSIS was noted from IL6-treated Min6 cells, no change in the total insulin content was noted. Coculture of Min6 cells with PSCs or their exposure to IL6 did not alter either the expression of β-cell-specific genes or that of miRNA-375. PSC-cocultured Min6 cells, in the presence of PLC-IP3 pathway inhibitors (U73122, Neomycin, and Xestospongin C), did not revoke the observed increase in GSIS. In conclusion, the obtained results indicate that augmented insulin secretion from Min6 cells in response to PSC secretions is independent of IL6-mediated PLC-IP3 pathway.
1 INTRODUCTION
Pancreatic stellate cells (PSCs) are well-known to secrete various profibrogenic1-3 and pro-inflammatory cytokines4-6 that contribute to a fibro-inflammatory milieu in the inflamed pancreas, as seen in disease conditions such as chronic pancreatitis (CP) and pancreatic cancer (PC). In addition to the inflammatory cells, PSC infiltration into the islets has recently been reported both in humans and in diabetic rodent models.7-11 The immunohistochemistry studies from our laboratory also demonstrated the infiltration of activated PSCs into the human islets from CP nondiabetic and diabetic pancreatic tissue,12 suggesting their possible contribution in establishing an inflammatory milieu and thus influencing the β-cell function. However, the underlying mechanism(s) demonstrating the influence of PSCs on β-cell function and their contribution in the pathogenesis of diabetes mellitus are unknown.
Activated PSCs are also reported to secrete interleukin 6 (IL6),13-16 activin-A,17-20 hepatocyte growth factor (HGF),21 and acetylcholine,22, 23 which are known to promote the insulin secretion from β-cells.24-26 Earlier studies also denoted that such an influence can be countered by cytokines such as interleukin-1β (IL1β),27, 28 tumor necrosis factor-α (TNFα), and interferon-γ (IFNγ)29-32 that impair insulin secretion from β-cells. Considering the ability of PSCs to secrete various cytokines and their identification within the islets in pancreatic diseases, we hypothesized that the activated PSCs may influence β-cell function. Hence, we studied the effect of in vitro-activated PSCs on Min6 cell insulin secretory function, expression of β-cell-specific genes, and miRNA-375.
2 MATERIALS AND METHODS
2.1 Chemicals
All the chemicals used in this study were purchased from Sigma-Aldrich, unless otherwise mentioned.
2.2 Isolation and characterization of pancreatic stellate cells from the C57BL6J mice
PSCs were isolated from the C57BL6J mice pancreas using the method of Apte et al,33 with minor modifications. In brief, the dissected mice pancreas was collected and cleaned in ice-cold phosphate-buffered saline (PBS), distended by injecting with enzyme solution containing Collagenase P (Roche, Germany) and DNase I (Roche) in Hank's balanced salt solution (HBSS) buffer, followed by incubation at 37°C for 5 minutes. The dissociated pancreatic tissue was then finely minced and incubated for 7 more minutes in a water bath. The obtained cell suspension was filtered through 150 μm nylon mesh and centrifuged at 450g for 10 minutes. The supernatant was discarded and the cell pellet was resuspended in HBSS solution containing 0.3% bovine serum albumin (BSA) and centrifuged for 10 minutes. The resulting cell pellet was once again resuspended in Nycodenz (Axis Shield, Norway) gradient solution, layered under HBSS containing 0.3% BSA solution and centrifuged at 1400g for 20 minutes at 4°C. The fuzzy band formed at the interphase of Nycodenz gradient and HBSS buffer was collected and resuspended in 10 mL of HBSS buffer and centrifuged at 450g for 10 minutes. The pellet containing primary PSCs were suspended in Iscove's modified Dulbecco's media (IMDM) (Himedia, India) containing 20% fetal bovine serum (FBS) (Himedia) along with 1% penicillin-streptomycin (Himedia) and incubated in a humidified chamber at 37°C with 5% CO2.
The isolated PSCs were characterized by immunostaining for glial fibrillary acidic protein (GFAP) (anti-GFAP mouse monoclonal at 1:20; 1 μg in 10 μL and goat polyclonal secondary antibody to mouse IgG-H&L-Texas Red at 1:500; Abcam) and α-smooth muscle actin (α-SMA) (anti-αSMA mouse monoclonal at 1:200; 0.1 μg in 100 μL and goat polyclonal secondary antibody to mouse IgG-H&L-DyLight 488 1:100; Abcam), a marker that is expressed by activated PSCs. Images were captured using fluorescent microscope (Olympus) and CARVII bioimager (BD Biosciences) using IP Lab software.
2.3 Culturing of Min6 cell line
Min6 cells were obtained from AddexBio, USA and regularly maintained in Dulbecco's modified Eagle medium (DMEM) (Himedia) containing 25 mM glucose, 0.05 mM β-mercaptoethanol, 1% penicillin and streptomycin, and supplemented with 15% FBS. The cells were maintained at 37°C in a humidified incubator with 5% CO2. All the experiments were conducted using Min6 cells between passages 14 and 25.
2.4 Indirect coculture of PSCs with Min6 cells using the Transwell inserts
Indirect coculture of PSCs and Min6 cells was carried out using the Transwell inserts (Corning, NY)8, 34 with minor modifications. On day 1, sub-confluent PSCs and Min6 cells were trypsinized and counted. PSCs suspended in complete IMDM were seeded on a Transwell polycarbonate membrane insert (pore size of 0.4 µm) at a seeding density of 0.1 × 104 cells/cm2 and placed in a culture well containing complete IMDM. Similarly, Min6 cells suspended in complete DMEM, were seeded in a 6- or 12-well plate at a seeding density of 0.3 × 105 cells/cm2. Both PSCs and Min6 cells were seeded at 1:30 ratio and the cells were left undisturbed for 24 hours in an incubator at 37°C with 5% CO2. On day 2, the spent medium was replaced with fresh complete medium for both cell types. Later, the Transwell inserts containing PSCs were placed on the wells seeded with Min6 cells. This experimental setup is herein referred to as “indirect coculture system” and wells containing only Min6 cells with Transwell inserts (monocultures) without PSCs were considered as controls. The incubation of monocultured and PSC-cocultured Min6 cells was continued for the next 72 hours and used in subsequent glucose-stimulated insulin secretion (GSIS) and quantitative polymerase chain reaction (qPCR) studies. Wherever required, PLC-IP3 inhibitors such as U73122 (Abcam) 2 µmol/L, Neomycin (Abcam) 1.5 mmol/L, and Xestospongin C (Abcam) 10 µmol/L were added to the coculture system after 24 hours and the incubation was continued for the next 72 hours as in coculture with these inhibitors. Further, the effect of these inhibitors on GSIS and the total insulin content was evaluated by enzyme-linked immunosorbent assay (ELISA).
2.5 Treatment of Min6 cells with PSC-conditioned medium
Sub-confluent PSCs at the second passage were trypsinized, counted, and seeded in T25 culture flasks containing complete IMDM at a density of 0.2 × 106 PSCs, per flask. At the end of 24 to 36 hours, the spent complete medium was replaced with fresh IMDM without FBS and the cells were incubated overnight, followed by replacing the medium with 5 mL of fresh IMDM supplemented with 1% FBS and incubation for a further period of 48 hours. Later, the PSC spent medium was collected and centrifuged at 450g for 10 minutes. The supernatant was filtered through 0.2 μm syringe filter and stored at −80°C until further use. Conditioned medium prepared using passage 3 PSCs was used to examine its influence on Min6 cells function. Min6 cells were then cultured for 48 hours in DMEM followed by replenishing with fresh DMEM containing conditioned medium (40%, v/v) and 1% FBS. Min6 cells were then incubated for a further period of 48 hours in a humidified incubator with 5% CO2 at 37°C to examine the influence of PSC-conditioned medium on insulin secretory responses and gene expression.
2.6 Glucose stimulated insulin secretion
At the end of the specified incubation periods, the culture medium was discarded and Min6 cells were washed gently using Krebs-Ringer bicarbonate-HEPES (KRBH) buffer of pH 7.4 containing (mmol/L): NaCl, 120; KCl, 5; CaCl2, 2.56; MgCl2, 1.1; NaHCO3, 25; Hepes, 10 (Himedia) and supplemented with 0.2% w/v BSA. Min6 cells were then preincubated in KRBH buffer containing 2.5 mM glucose for 30 minutes and the supernatants were discarded. Min6 cells were then incubated in KRBH buffer containing 2.5 mM glucose, followed by KRBH buffer containing 25 mM glucose and samples were collected at 10, 20, and 60 minutes, respectively. Insulin secretion by these cells was measured in the supernatants and insulin content in cell lysates was estimated employing ELISA (Mercodia, Sweden) as per manufacturer's instructions and measurement at 450 nm using microplate reader (Bio-Rad 680, Japan). Insulin secretion in response to glucose stimulation and total insulin contents were expressed in relation to per million cells in coculture experiments and to total protein content in the experiments involving Min6 cell treatment with IL6.
2.7 RNA extraction, Reverse-transcription PCR and qPCR
The total RNA was extracted from Min6 cells using TRIzol reagent (Ambion). DNA contamination in total RNA was eliminated by treating the isolates with 5 U/µL DNase I (Takara, Japan) for 30 minutes at 37°C, followed by its inactivation at 80°C for 2 minutes. After ascertaining concentration and purity of the isolated RNA using ND-100 spectrophotometer, 1 µg of total RNA was subjected to complementary DNA (cDNA) synthesis employing 50 µM random hexamers (Invitrogen) and 10 mM dNTPs Mix (Thermo Fisher Scientific) in a total reaction volume of 20 μL containing 40 U/µL RNaseOUT (Invitrogen) to inhibit the RNase activity. The reaction mixture was incubated at 55°C for 5 minutes, and reverse-transcription was carried out using 200 U/µL Superscript IV (Invitrogen) for 30 minutes at 37°C. The cDNA thus prepared was then stored at −20°C until use. All the primers are designed using Integrated DNA Technologies online real-time PCR tool. All the primers were used at a working concentration of 10 pmol. The primer sequences and their efficiencies and amplicon sizes are listed in Table 1. The images of qPCR melt curves and gel picture showing single PCR product were included in Figure S4. In total, 0.5 μL of cDNA was added in a reaction volume of 10 μL to study the relative expression of β-cell-specific genes by using the SYBR green (Applied Biosystems, UK) chemistry on StepOne Real-Time PCR System (Applied Biosystems). β-Actin was used as an endogenous control to normalize the target gene expression levels. The differential expression of target genes was calculated using method and represented in the form of fold change.
| Gene | Transcript ID | Forward and reverse primer sequences | Amplicon size, bp | Primer efficiency |
|---|---|---|---|---|
| Ins | NM_008386 | 5-GCCCTTAGTGACCAGCTATAATC-3 | 154 | 89.41 |
| 5-GGACCACAAAGATGCTGTTTG-3 | ||||
| Pdx-1 | NM_008814 | 5-GAAATCCACCAAAGCTCACG-3 | 190 | 87.81 |
| 5-CAAGTTCAACATCACTGCCAG-3 | ||||
| Rfx6 | NM_001159389 | 5-CGGTGCATTCTTTATGCTCA-3 | 219 | 90.51 |
| 5-TGTCAAGCCCTTTCCAGAAT-3 | ||||
| MafA | NM_194350 | 5-GAGGTCATCCGACTGAAACAG-3 | 203 | 103.05 |
| 5-GCCAACTTCTCGTATTTCTCCT-3 | ||||
| NeuroD1 | NM_010894 | 5-CCAGGGTTATGAGATCGTCAC-3 | 171 | 85.01 |
| 5-TTCTTGTCTGCCTCGTGTTC-5 | ||||
| Nkx2-2 | NM_001077632 | 5-TTCCATAACCATCGCTACAAG-3 | 236 | 81.31 |
| 5-TTGGCATTGTGGTCCTACTG-3 | ||||
| Mtpn | NM_008098 | AAAACGGAGACTTGGATGAGG | 247 | 89.71 |
| TCAGCACCCTTTGACAGAAG | ||||
| β-Actin | NM_007393 | 5-CATCCGTAAAGACCTCTATGCC-3 | 231 | 84.47 |
| 5-GACTCATCGTACTCCTGCTTG-3 |
2.8 MicroRNA isolation, RT-PCR, and qPCR
Total RNA-enriched with miRNA was isolated from Min6 cells using miRNeasy mini Kit (Qiagen, Germany). The quality and quantity were assessed using ND-100 spectrophotometer. RT-PCR for target miRNAs was performed according to the protocol given with TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Lithuania), using the stem-loop primers from TaqMan microRNA assays (miRNA-375 Assay ID 000564, U6 snRNA Assay ID 001712, Applied Biosystems) in a total volume of 15 µL per reaction. qPCR was performed on StepOne Real-Time PCR System (Applied Biosystems) using respective 20× TaqMan probes and TaqMan Universal PCR Master Mix, no AmpErase UNG protocol (Applied Biosystems) in a total reaction volume of 20 µL. U6 snRNA and Sno234 were used as endogenous controls to normalize the expression levels of miRNA-375 in the respective studies and the relative expression was calculated using method.
2.9 Quantification of cytokines in cell culture supernatants
Cytokines such as IL2, IL4, IL6, IL17A, IL10, IFNγ, and TNFα were quantified in the cell culture supernatants using Cytometric Bead Array (Mouse Th1/Th2/Th17 Cytokine Kit; BD Biosciences). Serially diluted standards and capture bead mixture for the cytokines were prepared as per the manufacturer's protocol. In brief, 50 µL of the capture bead mixture was added to 50 µL of serially diluted standards and the samples under study, followed by the addition of PE detection reagent (50 µL) to all the assay tubes. After a 2-hour incubation period at room temperature in the dark, 1 mL of wash buffer was added to all the assay tubes and the contents were centrifuged at 200g for 5 minutes. After discarding the supernatant, 300 µL of wash buffer was added to each assay tube and bead pellet was resuspended. The samples were acquired on BD FACS Aria II machine and the data were analyzed using FACS Diva software.
2.10 Treatment of Min6 cells with IL6
Min6 cells were seeded at a density of 0.3 to 0.4 × 105 cells/cm2 in complete DMEM and incubated for 24 to 48 hours. The spent medium was replaced with DMEM supplemented with 1% FBS containing IL6 (Peprotech, Israel) at 50, 250, and 1000 pg/mL concentrations. Min6 cells in DMEM supplemented with 1% FBS and without IL6 were used as controls. The cells were incubated in a CO2 incubator at 37°C for 48 hours. These cells were used to study the influence of IL6 on GSIS and relative expression of β-cell-specific genes and miRNA-375 as described above.
2.11 Annexin V and propidium iodide staining
Min6 cells were subjected to PSC coculture and its conditioned medium as well as IL6 treatment accordingly as described in the methods. After the completion of incubation period, the cells were washed three times in PBS, trypsinized, and centrifuged at 130g for 5 minutes. The Min6 cells in 15 mL falcon tubes were resuspended in 5 mL of complete DMEM and incubated in a CO2 incubator for 30 to 40 minutes. Then, the cell number was adjusted to 0.1 × 106 cells/100 μL of 1× annexin V binding buffer. To this cell suspension, 5 μL of fluorescein isothiocyanate (FITC)-conjugated annexin V (Invitrogen) and 5 μL of propidium iodide (Invitrogen) were added and incubated for 10 to 15 minutes at room temperature in the dark. These cells were then suspended in 400 μL of 1× annexin V binding buffer. Annexin V-FITC binding was detected by using FITC signal detector and propidium iodide staining by the phycoerythrin emission signal detector on BD FACS ARIA II and the data were analyzed.
2.12 Detection of cleaved caspase-3 by flow cytometry
Cleaved caspase-3 was detected in Min6 cells from the control and experimental groups following the method described by Crowley and Waterhouse.35 In brief, Min6 cells were harvested and washed twice with ice-cold PBS at 130g for 5 minutes. Min6 cells were resuspended in ice-cold 4% paraformaldehyde and incubated for 20 minutes at 4°C and washed twice with ice-cold PBS. Later, the cells were washed in 1 mL of IFA-Tx buffer for 5 minutes at 130g. Min6 cells were then incubated in 150 µL of primary antibody master mix (Rabbit polyclonal to caspase-3, 1:500 dilution; 1 μg in 500 μL; Abcam) and incubated for 1 hour at 4°C. Min6 cells were then washed in IFA-Tx buffer for three times and incubated in 150 µL of secondary antibody (Goat Antirabbit IgG-H&L -Alexa Fluor 488, 1:1000 dilution; Abcam) for 1 hour at 4°C. After the completion of the incubation period, Min6 cells were washed in 1 mL of PBS for three times and resuspended in 150 μL of PBS. The samples were run on BD FACS ARIA II using 488 nm laser and the data acquired for 10 000 events.
2.13 Statistical analysis
P values were calculated using Student's t test and/or analysis of variance. Data are represented as mean ± SEM unless otherwise mentioned. P ≤ .05 was considered to be significant.
3 RESULTS
3.1 Propagation and characterization of PSCs
PSC isolation procedure from mouse pancreas regularly yielded PSCs of greater than 95% purity and attained their typical morphology within 6 to 10 hour after isolation, showing dense lipid droplets surrounding the nucleus (Figure 1A). The presence of dense lipid droplets was observed for 48 hour and gradual transformation of PSCs into myofibroblast phenotype along with the disappearance of lipid droplets was observed after passage 1. Isolated PSCs were characterized by immunostaining for GFAP and α-SMA (Figure 1B and 1C). The images of monocultured and PSC-cocultured Min6 cells, along with PSC CM-treated Min6 cells were included in Figure S3.
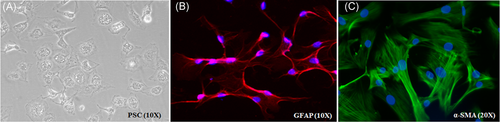
Characterization of isolated PSCs. A, PSCs showing typical polygonal shape with translucent lipid droplets in the cytoplasm after 24 hours of isolation (×10). B, PSCs immunostained for GFAP (red) (×10), and (C) α-SMA (green), a marker representing the activated phenotype of PSCs (×20). Nuclei in blue color are stained with DAPI. α-SMA, α-smooth muscle actin; DAPI, 4′,6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein; PSC, pancreatic stellate cell
3.2 Activated PSC secretions increased GSIS from Min6 cells
Min6 cells cocultured with activated PSCs showed a significant increase in insulin secretion in response to high glucose challenge as compared with monocultured Min6 cells at 20 minutes (coculture [CC]: 9.80 ± 0.71; monoculture [MC]: 5.20 ± 0.36 pmol/106 cells; P ≤ .05) and 60 minutes (CC: 10.08 ± 0.97; MC: 5.47 ± 0.52 pmol/106 cells; P ≤ .05), respectively, when compared with insulin secretion at 10-minute time point (CC: 1.55 ± 0.33; MC: 1.65 ± 0.21 pmol/106 cells; P = ns). However, no significant variation in the amount of insulin secreted was noted between monoculture and cocultures at 20- and 60-minute time points (Figure 2A). Similarly, Min6 cells cultured in PSC-conditioned medium also demonstrated increased insulin secretory response at 20 minutes (PSC CM: 9.01 ± 0.5; control [Con]: 5.18 ± 0.38 pmol/106 cells; P≤ .05) and 60 minutes (PSC CM: 10.37 ± 1.2; Con: 5.29 ± 0.25 pmol/106 cells; P ≤ .05), respectively, when compared with insulin secretion at 10-minute time point (PSC CM: 1.69 ± 0.37; Con: 1.76 ± 0.51 pmol/106 cells; P = ns). Similar to PSC coculture experiment, control and PSC-conditioned medium-treated Min6 cells did not show any significant difference in the amount of insulin secreted at 20- and 60-minute time points (Figure 2D).
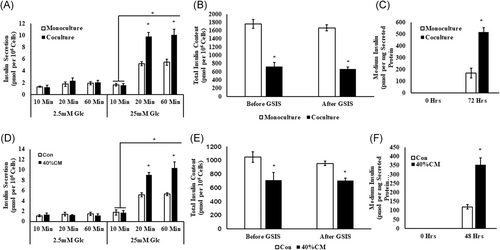
Influence of activated PSCs (A-C) and its conditioned medium (D-F) on insulin and total insulin content in Min6 cells: A and D, Increased GSIS, (B and E) decreased total insulin content, and (C and F) elevation in medium insulin levels were observed in PSC-cocultured and conditioned medium-treated Min6 cells, when compared with control Min6 cells. Insulin secreted and total insulin contents and medium insulin levels were normalized to per million cells. The Kruskal-Wallis test followed by post hoc analysis for comparison between multiple groups was used to analyze the data presented in (A) and (D) using Med Calc application and ANOVA for the data presented in (B, C, E, and F). Results are presented as mean ± SEM from n = 3 independent experiments and the differences were considered significant when *P ≤ .05. CM, conditioned medium; Con, control; Glc, glucose; GSIS, glucose-stimulated insulin secretion
3.3 Total insulin contents in PSC-cocultured Min6 cells before and after GSIS
A significant decrease in the total insulin content was noticed in Min6 cells cocultured with PSCs (CC: 725.72 ± 76.51; MC: 1762.59 ± 110.72 pmol/106 cells; P ≤ .05) in comparison with Min6 cells cultured without PSCs (Figure 2B), after conducting the GSIS. To understand whether the decrease in the total insulin content noted in the PSC-cocultured Min6 cells is the outcome of increased GSIS, the total insulin content was quantified in Min6 cell before performing the GSIS. As shown in Figure 2B, a significant variation in the amount of total insulin content (CC: 666.11 ± 50.76; MC: 1665.53 ± 100.50 pmol/106 cells; P ≤ .05) was noted in monoculture when compared with coculture. However, no statistical significance was attained in the amount of insulin content retained within the groups, before and after GSIS.
Similar to PSC-cocultured Min6 cells as shown in Figure 2E, PSC-conditioned medium-treated Min6 cells also showed a significant variation in the total insulin content measured before (PSC CM: 706 ± 31.55; Con: 954.91 ± 114.52 pmol/106 cells; P ≤ .05) and after conducting GSIS (PSC CM: 708 ± 33.01; Con: 1049.62 ± 77.37 pmol/106 cells; P ≤ .05), when compared with control Min6 cells. Although significant change in the total insulin content was noted between the control and PSC-conditioned medium-treated groups, no substantial variation was noted in the amount of insulin retained within the same groups before and after the GSIS, suggesting no contribution of increased GSIS on decreased total insulin content in PSC-cocultured and conditioned medium-treated Min6 cells.
3.4 Increase in insulin levels in the coculture medium
As shown in Figure 2C and 2F, quantification of medium insulin levels presented a raise in the their levels, both in PSC-cocultured (CC: 516.5 ± 37.49; MC: 169.67 ± 41.24 pmol/106 cells; P ≤ .003) and conditioned medium (PSC CM: 354.30 ± 37.35; Con: 120.03 ± 12.37 pmol/106 cells; P ≤ .05)-treated Min6 cell group, when compared with control Min6 cells. These observations suggest that the noted decrease in the total insulin content may rather be owing to the elevation of insulin levels in the medium in PSC-cocultured and conditioned medium-treated Min6 cells, but not owing to increased insulin secretion during GSIS.
3.5 Activated PSCs do not influence the expression of β-cell-specific genes
Although PSCs-cocultured Min6 cells showed a mild increase in the expression of Ins (0.61 ± 0.57 folds; P ≤ .39), NeuroD1 (1.15 ± 0.69 folds; P ≤ .23), and Nkx2-2 (0.38 ± 0.30 folds; P ≤ .39) genes, the observed changes were without statistical significance. Similarly, the expression of transcription factors such as Pdx1 (1.00 ± 0.30 folds; P ≤ .98), MafA (0.12 ± 0.04 folds; P ≤ .11), and Rfx6 (0.11 ± 0.05 folds; P ≤ .18) was unaltered in Min6 cells cocultured with PSCs as compared with those cultured in the absence of PSCs (Figure 3A). As shown in Figure 3B, similar results were noticed even in Min6 cells cultured in PSC-conditioned medium.
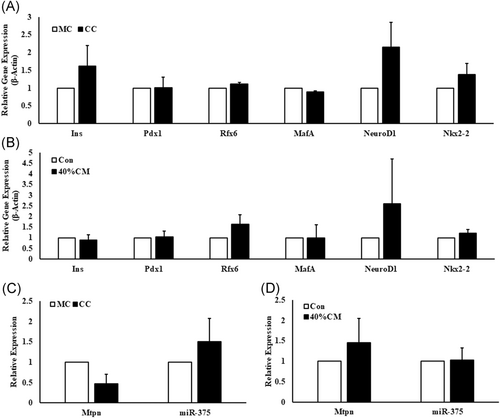
Relative expression of β-cell-specific genes in PSC-cocultured and conditioned medium-treated Min6 cells. The β-cell genes such as Ins, Pdx1, Nkx2-2, MafA, NeuroD1, and Rfx6 did not show any significant fold change in their expression neither in (A) PSC-cocultured nor in (B) PSC-conditioned medium-treated Min6 cells. β-Actin was used as endogenous control to normalize the expression of the target genes. (C) Mtpn and miR-375 expression levels showed no significant change in both PSC-cocultured Min6 cells and (D) PSC-conditioned medium-treated Min6 cells. The miR-375 expression was normalized to U6 snRNA expression. Unpaired Student's t test was used to calculate the significance using Microsoft Excel 2016 application. Data are represented as mean ± SEM from n = 3 independent experiments. CC, coculture; CM, PSC-conditioned medium; Con, control; MC, monoculture; miR-375, microRNA-375; Mtpn, myotrophin; PSC, pancreatic stellate cell. P = ns compared with MC and/or Con Min6 cells
3.6 Activated PSCs did not alter the expression of miRNA-375
In view of earlier observations indicating negative regulation of GSIS by miRNA-375, by modulating myotrophin expression,36 we investigated whether the inhibition of miRNA-375 expression is contributing to increased GSIS. The expression levels of mature miRNA-375 was relatively unaffected when Min6 cells were cocultured with PSCs or when they were cultured in PSC-conditioned medium, the values being 0.51±0.56-folds (P ≤ .45) and 1.02±0.30 folds (P ≤ .94) respectively. Myotrophin expression was found to be decreased (0.53±0.22 fold; P ≤ .14) in PSC-cocultured Min6 cells, while its expression was increased (0.54±0.59-fold; P ≤ .51) in conditioned medium-treated Min6 cells when compared with Min6 cells alone (Figure 3C and 3D). However, no statistical significance was achieved in neither of the experimental groups.
3.7 Activated PSCs secrete IL6
Analysis of the cell culture supernatants obtained from monoculture and coculture for cytokines (IFNγ, IL6, TNFα, IL2, IL4, IL10, and IL17A) identified the presence of only IL6 (261 ± 33.95 pg/mL; P ≤ .005) in the coculture system. Similarly, only IL6 (456 ± 68.66 pg/mL; P ≤ .003) was detected in the PSC-conditioned medium (Figure 4A and 4B), among the seven different cytokines profiled.
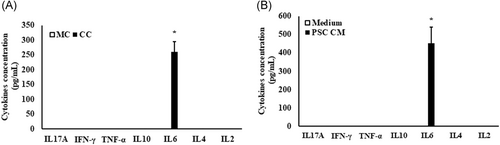
Quantification of the cytokines in cell culture supernatants. A, Cell culture supernatants from monocultured and PSC-cocultured Min6 cells and (B) PSC-conditioned medium were collected and used for qualitative and quantitative detection of cytokines such as IL17A, IFNγ, TNFα, IL10, IL6, IL4, and IL2. Except IL6 from coculture and PSC-conditioned medium, none of the cytokines tested were detected. Data are represented as mean ± SD from n = 3 independent experiments and each sample was analyzed in duplicates. Unpaired Student's t test was used to calculate the significance, using Microsoft Excel 2016. *P ≤ .005 compared with monoculture and/or control medium. CC, coculture; CM, PSC-conditioned medium; MC, monoculture
3.8 IL6 increased the GSIS from Min6 cells without altering the total insulin content
Significant increase in the insulin secretion by Min6 cells was noticed when they were pretreated with IL6. In comparison with untreated controls (7.16 ± 0.49 pmol/mg protein), pretreatment of Min6 cells with 250 and 1000 pg/mL of IL6, resulted in increase of GSIS by 13.11 ± 0.43 and 15.2 ± 0.26 pmol/mg protein, respectively (Figure 5A). Unlike PSC-cocultured Min6 cells, IL6-treated Min6 cells showed no change in the total insulin content (IL6 control: 792.02 ± 16.16; 50 pg/mL: 796.95 ± 31.18; 250 pg/mL: 766.09 ± 29.09; 1000 pg/mL: 762.08 ± 56.92 pmol/mg protein; P = ns), as shown in Figure 5B. In addition to this, quantification of medium insulin levels in IL6-treated Min6 cells at 0 and 48 hours also did not show significant variation (Figure S1).
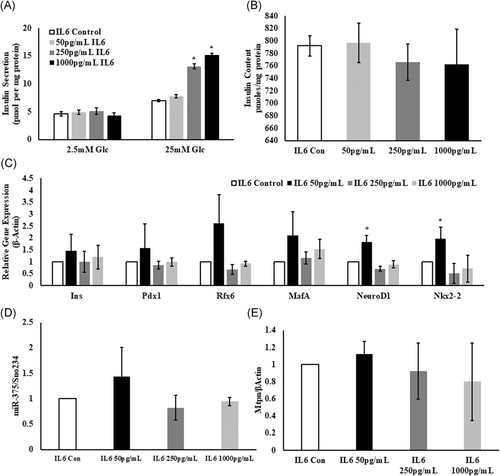
Influence of IL6 on Min6 cell function. A, Similar to PSC-cocultured Min6 cells, enhanced GSIS was noticed from Min6 cells incubated with 250 and 1000 pg/mL IL6 with no significant change in insulin secretion at 50 pg/mL IL6 and (B) unlike to PSC-cocultured Min6 cells, total insulin content did not vary in IL6-treated Min6 cells when compared with control Min6 cells. The amount of insulin secreted and total insulin content were corrected to total protein. Data are represented as mean ± SEM from n = 3 independent experiments and each sample was analyzed in duplicates. *P ≤ .01 compared with control Min6 cells. C, IL6-treated Min6 cells showing differential expression of β-cell-specific genes with no significance, except for NeuroD1 and Nkx2-2 at 50 pg/mL IL6 (n = 3 independent experiments), *P ≤ .01. D, Differential expression of miR-375 (n = 3) and (E) Mtpn (n = 3) in IL6-treated Min6 cells showing no statistical significance. P = ns compared with control Min6 cells. Vasserstats one-way analysis of variance was used to calculate the P value. Con, control; Glc, glucose; Mtpn, myotrophin
3.9 IL6 did not influence the expression of β-cell-specific genes and miR-375
Differential expression of Ins, Pdx1, Rfx6, NeuroD1, MafA, and Nkx2-2 was noticed in Min6 cells treated with 50, 250, 1000 pg/mL IL6. However, as shown in Figure 5C, only NeuroD1 and Nkx2-2 showed a statistically significant fold change in response to the addition of IL6 (50 pg/mL) when compared with Min6 cells, and that were not exposed to IL6. Similarly, although miR-375 (Figure 5D) and Mtpn (Figure 5E) showed a marginal change in their expression levels, the fold difference observed between the control and IL6-treated Min6 cells did not reach the statistical significance. The fold changes observed in response to the addition of IL6 in the culture medium are shown in Table 2.
| Gene | IL6 control | 50 pg/mL | 250 pg/mL | 1000 pg/mL |
|---|---|---|---|---|
| Ins | 1 | 1.46 ± 0.69 | 1.00 ± 0.44 | 1.19 ± 0.5 |
| Pdx1 | 1 | 1.57 ± 1 | 0.85 ± 0.16 | 0.98 ± 0.18 |
| Rfx6 | 1 | 2.61 ± 1.27 | 0.67 ± 0.21 | 0.92 ± 0.1 |
| MafA | 1 | 2.1 ± 0.99 | 1.15 ± 0.25 | 1.5 ± 0.4 |
| NeuroD1 | 1 | 1.83 ± 0.27 | 0.70 ± 0.10 | 0.88 ± 0.15 |
| Nkx2-2 | 1 | 1.96 ± 0.48 | 0.51 ± 0.42 | 0.71 ± 0.57 |
| Mtpn | 1 | 1.12 ± 0.14 | 0.92 ± 0.32 | 0.79 ± 0.45 |
3.10 Activated PSCs and IL6 do not induce apoptosis in Min6 cells
In addition to the in vitro-activated PSCs and its conditioned medium, the influence of IL6 on Min6 cell viability was evaluated by annexin V/propidium iodide staining, to distinguish the live (AnnexinV−/PI−), early apoptotic (AnnexinV+/PI−), and late apoptotic (AnnexinV+/PI+) cells. The percentage of viable, along with early and late apoptotic cells were noticed to be similar in different experimental groups including PSCs-cocultured (Figure 6A-C), conditioned medium (Figure 6D-F), and IL6-treated Min6 cells (Figure 6G-K). These results suggest that neither the in vitro-activated PSCs nor IL6 contribute to the Min6 cell death under in vitro conditions. Similarly, the percentage of cleaved caspase-3 Min6 cells was found to be similar in both PSC coculture (Figure 7A-C) and CM-treated (Figure 7D-F) groups when compared with control groups. Likewise, no significant variation in the percentage of cleaved caspase-3 Min6 cells was observed even in IL6-treated Min6 cells (Figure 7G and 7H) in comparison with control Min6 cells. The percentage of viable, early, and late apoptotic cells along with cleaved caspase-3-positive population are tabulated in Table 3.
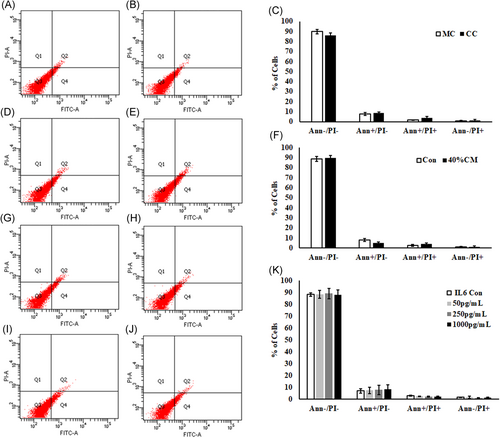
Analysis of Annexin V/PI staining in Min6 cells. Min6 cells cocultured with PSCs for 72 hours were subjected to Annexin V/PI staining and analyzed by FACS. A and B, Representative scatter plots were shown for monocultured and PSC-cocultured Min6 cells (C) 2D columns representing the percentage of viable, early, and late apoptotic Min6 cells from monoculture and coculture. D and E, Illustrative scatter plots showing the distribution of control and PSC-conditioned medium-treated Min6 cells population after staining with Annexin V/PI in four quadrants. F, 2D histograms showing the percentage of live, early, and late apoptotic Min6 cell population. Min6 cells incubated with 50, 250, and 1000 pg/mL concentration of IL6 for 48 , stained for Annexin V/PI and analyzed by flow cytometry. G-J, Representative scatter plots showing the distribution of Min6 cell population treated with IL6 in four different quadrants after staining with Annexin V/PI. K, Histograms showing the percentage of viable, early, and late apoptotic cells Min6 cell population after treatment with IL6. Data represent mean ± SD from three independent Min6 cell preparations. P = ns vs control Min6 cells without IL6 treatment. Unpaired Student's t test was used to calculate the significance between two groups and analysis of variance in IL6-treated Min6 cells. PI, propidium iodide
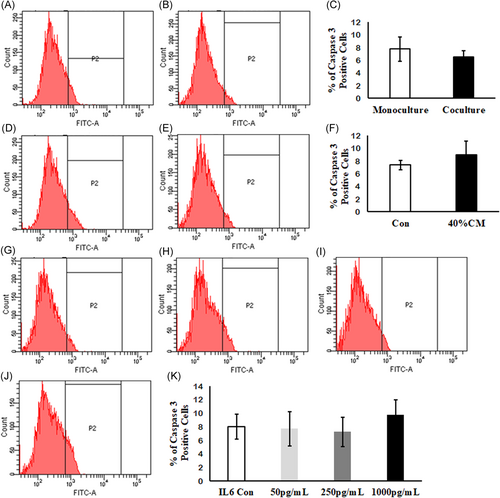
Detection of cleaved caspase-3 in Min6 cells by flow cytometry. Representative flow cytometry histograms for PSC-cocultured (A and B), conditioned medium (D and E), and IL6-treated Min6 cells (G-J). Two-dimensional bars showing no significant variation in percentage of cleaved caspase-3-positive cells in Min6 cells treated with (C) PSCs, (F) conditioned medium, and (K) IL6. Data represent mean ± SD from three independent Min6 cell preparations. P = ns vs control Min6 cells without IL6 treatment. Unpaired Student's t test was used to calculate the significance between two groups and one-way analysis of variance in IL6-treated Min6 cells
| Experiment condition | Ann−/PI− | Ann+/PI− | Ann+/PI− | Ann−/PI+ | Caspase 3 |
|---|---|---|---|---|---|
| Monoculture | 89.73 ± 2.14 | 7.83 ± 1.46 | 1.6 ± 0.30 | 0.83 ± 0.42 | 7.78 ± 1.9 |
| Coculture | 85.93 ± 2.30 | 8.7 ± 1.06 | 3.83 ± 1.19 | 1.53 ± 0.84 | 6.5 ± 0.96 |
| Control | 88.76 ± 2.25 | 7.96 ± 1.28 | 2.43 ± 1.00 | 0.83 ± 0.83 | 7.36 ± 0.72 |
| 40% CM | 89.9 ± 2.32 | 4.96 ± 0.89 | 3.86 ± 1.13 | 1.26 ± 0.75 | 9.0 ± 2.12 |
| IL6 Con | 88.13 ± 1.57 | 7.16 ± 1.88 | 3.06 ± 0.33 | 1.63 ± 0.08 | 8 ± 1.83 |
| 50 pg/mL IL6 | 88.23 ± 3.26 | 7.46 ± 2.68 | 2.56 ± 0.17 | 1.73 ± 0.98 | 7.7 ± 2.55 |
| 250 pg/mL IL6 | 89.03 ± 4.43 | 7.86 ± 3.82 | 2.1 ± 0.45 | 1 ± 0.52 | 7.24 ± 2.15 |
| 1000 pg/mL IL6 | 87.83 ± 4.12 | 8.33 ± 3.9 | 2.26 ± 0.47 | 1.56 ± 0.36 | 9.76 ± 2.52 |
- Abbreviations: CM, PSC-conditioned medium; Con, control; PI, propidium iodide.
3.11 Inhibition of IL6-mediated PLC-IP3 pathway did not influence the GSIS and total insulin content in PSC-cocultured Min6 cells
To verify the involvement of PLC-IP3 pathway in the IL6-induced augmentation of GSIS by Min6 cells cocultured with PSCs, the influence of known inhibitors of the PLC-IP3 pathway was examined. As shown in the Figure S2, GSIS was attenuated from Min6 cells preincubated with 1000 pg/mL IL6 along with the PLC-IP3 pathway inhibitors, for a period of 48 hours. Further, inclusion of U73122 (a PLC inhibitor; 2 μmol/L) or Neomycin (a PIP2 inhibitor; 1.5 mmol/L) or Xestospongin C (an IP3 inhibitor; 10 μmol/L) did not cause any significant change in the amount of insulin secreted from Min6 cells cocultured with PSCs (Figure 8). Insulin secretion from Min6 cells cultured in absence and in presence of PSCs and in response to inhibitors of PLC-IP3 pathway are shown in Table 4.
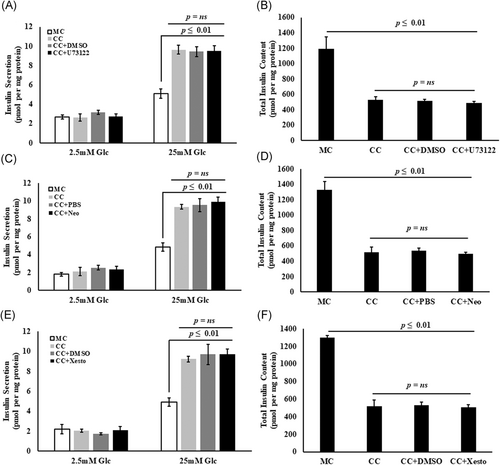
Abrogation of PLC-IP3 pathway in the coculture system does not influence the increased insulin secretion from PSC-cocultured Min6 cells. A, C, and E, PSC-cocultured Min6 cells showing increased insulin secretion upon high glucose stimulation compared with monocultures. The presence of PLC-IP3 inhibitors such as 2 μmol/L U73122, a PLC inhibitor and/or 1.5 mmol/L Neomycin, a PIP2 inhibitor and/or 10 μmol/L Xestospongin C, an IP3 inhibitor did not show any significant reduction in the amount of insulin secreted upon high glucose stimulation compared with PSC-cocultured Min6 cells without inhibitors (*P ≤ .01). B, D, and F, PSC-cocultured Min6 cells showing a significant decrease in the total insulin content. A decrease in the total insulin content was even noted in the PSC-cocultured Min6 cells incubated with PLC-IP3 inhibitors (*P ≤ .01), suggesting no possible influence of PSC-secreted IL6-mediated PLC-IP3 pathway on Min6 cells when cocultured with PSCs. Data are represented as mean ± SEM from n = 3 independent experiments and P value was determined using Vasserstats one-way analysis of variance. DMSO and PBS were used as vehicle in the respective experiments. CC, coculture; DMSO, dimethyl sulfoxide; Glc, glucose; MC, monoculture; Neo, neomycin; Xesto, Xestospongin C
| Basal glucose | High glucose | Total insulin content | |
|---|---|---|---|
| GSIS responses observed in PSC CC Min6 cells with PLC inhibitor | |||
| MC | 2.7 ± 0.20 | 5.1 ± 0.36 | 1189.33 ± 160.36 |
| CC | 2.63 ± 0.46 | 9.63 ± 0.44 | 529.66 ± 39.25 |
| CC + DMSO | 3.16 ± 0.23 | 9.43 ± 0.49 | 515.33 ± 23.05 |
| CC + U73122 | 2.73 ± 0.26 | 9.5 ± 0.56 | 486.66 ± 20.66 |
| GSIS responses observed in PSC CC Min6 cells with PIP2 inhibitor | |||
| MC | 1.81 ± 1.90 | 4.86 ± 0.48 | 1325.33 ± 110.76 |
| CC | 2.13 ± 0.49 | 9.36 ± 0.26 | 517.66 ± 67.18 |
| CC + PBS | 2.53 ± 0.29 | 9.56 ± 0.72 | 538.32 ± 67.11 |
| CC + Neomycin | 2.36 ± 0.33 | 9.93 ± 0.52 | 496.66 ± 18.54 |
| GSIS responses observed in PSC CC Min6 cells with IP3 inhibitor | |||
| MC | 2.23 ± 0.47 | 4.91 ± 0.15 | 1301.66 ± 20.41 |
| CC | 2.06 ± 0.41 | 9.2 ± 0.27 | 516.33 ± 74.53 |
| CC + DMSO | 1.73 ± 0.12 | 9.7 ± 1.00 | 529.67 ± 37.54 |
| CC + Xestospongin C | 2.1 ± 0.36 | 9.73 ± 0.49 | 509.54 ± 29.86 |
- Abbreviations: CC, coculture; DMSO, dimethyl sulfoxide; GSIS, glucose-stimulated insulin secretion; MC, monoculture; PBS, phosphate-buffered saline; PSC, pancreatic stellate cell.
4 DISCUSSION
PSC infiltration into islets was reported in human CP, PC, and type 2 diabetic pancreatic tissues from rodents. Although, few in vivo studies9, 11 conducted in type 2 diabetic animal models demonstrated the contribution of PSCs to islet fibrosis, results on insulin secretory response from in vitro-activated PSC and islet/β-cell line coculture studies are not consistent.8, 37, 38 Hence, we conducted this study with an aim to understand the influence of activated PSCs on insulin secretion apart from studying the expression of β-cell-specific genes and miRNA-375 in a Min6 cell line.
In our study, we have identified increased insulin secretion with a concomitant decrease in the total insulin content in PSC-cocultured Min6 cells. Our results are in accord with an earlier study, which also demonstrated increased insulin secretion from PSC-cocultured islets.37 Based on these observations, it is surmised that PSCs may secrete certain factors to promote insulin secretion by Min6 cells at high glucose concentrations. Next, we tried to understand whether the observed increased insulin secretion is an outcome of the cross-talk between the Min6 cells and PSCs or whether it can be ascribed to factors secreted by PSCs. To address this question, Min6 cells were cultured in a PSC-conditioned medium and GSIS responses were studied. Similar to PSC-cocultured Min6 cells, PSC-conditioned medium-treated Min6 cells also showed similar experimental results, indicating that the results observed in the coculture experiment are rather due to the influence of PSC-secreted factors but not due to the cross-talk between the PSCs and Min6 cells. Interestingly, earlier investigations have shown similar kind of observations when islets were cocultured with bone marrow-derived mesenchymal stem cells (MSCs),21, 39-41 suggesting the participation of common factor(s) arising from PSCs and MSCs in promoting insulin secretion from β cells. In contrast to these results, a couple of studies conducted using RINm-5F8 and INS-1 cell line38 of rat origin showed decreased insulin secretion upon culturing with the activated PSCs and its conditioned medium, respectively. However, it is well-known that the RINm-5F cell line do not respond to high glucose stimulation in a consistent manner.42-44
An in vivo study by Lee et al11 reported a positive correlation of glucose intolerance with the percentage of the activated PSCs and islet fibrosis in OLETF animal model of type 2 diabetes mellitus. Although treating these animals with antifibrotic agent pirfenidone reduced the fibrosis, no beneficial effect in improving the glucose tolerance was observed. These in vivo experimental study results suggested that the activation of PSCs may cause islet fibrosis, but may not essentially contribute to progressive beta cell failure in type 2 diabetes. Although Lee et al study evaluated the glucose tolerance with relevance to islet fibrosis, insulin secretory responses in these animal models were not determined, which is the limitation of this study. The results from this study cannot be compared withour in vitro study. The basic idea of our study was to understand the in vitro effects of activated PSCs on Min6 cells with an emphasis on GSIS and gene expression, while Lee et al's study focused on glycaemic control.
Further, we examined the mRNA levels of insulin and its transcription factors to understand whether the decreased total insulin content detected in the cocultured Min6 cells was the outcome of the dysregulation of these genes. Interestingly, we did not identify any significant difference in mRNA levels related to insulin and other β-cell-specific genes in PSC cocultured Min6 cells. This result indicated that the observed decrease in total insulin content in the coculture may not essentially be due to the synthesis defect, but could be due to (a) continuous stimulation of insulin secretion from islets and Min6 cells by the PSC-secreted factors as reported earlier37 and/or (b) feedback inhibition of insulin synthesis due to the accumulated insulin in the medium.
One of the main events during β-cell insulin secretory response is the elevation of the cAMP levels, leading to insulin granule exocytosis.45, 46 Increased cAMP levels are known to repress miRNA-375 expression, which is mediated by PKA-dependent pathway.47 MicroRNA-375 is known to inhibit insulin exocytosis by posttranscriptional regulation of myotrophin, which is independent of glucose metabolism and [Ca2+]i levels.36 In view of these reports, we speculated that the PSC-secreted factors might be repressing the miRNA-375 expression and thus promote the myotrophin expression, leading to enhanced insulin secretion. Surprisingly, we did not identify any significant change in miRNA-375 or in myotrophin expression levels in PSC-cocultured and conditioned medium-treated Min6 cells. Based on the observed results, it is held that enhanced insulin secretion by Min6 cells may not be regulated through a mechanism driven by miRNA-375.
Cytokine analysis of cell culture supernatants obtained both from monoculture and coculture along with PSC-conditioned medium revealed that among the seven common cytokines examined (IFNγ, IL6, TNFα, IL2, IL4, IL10, and IL17A), only IL6 was detected in the coculture and PSC-conditioned medium samples and none were detected in Min6 monocultures (Figure 4A and 4B). These results suggest that the in vitro-activated PSCs secrete IL6, which was previously reported to enhance the GSIS.13-16 As IL6 has previously been reported to increase the GSIS via PLC-IP3 pathway,15 we studied its influence on GSIS by Min6 cells. Incubation of Min6 cells with IL6 increased the GSIS, with no apparent change in the relative gene expression and miR-375 as observed in the PSC-cocultured Min6 cells. These results made us to assume that PSC-secreted IL6 may have an important role in augmenting GSIS from Min6 cells.
On the basis of the observed results, we assumed that enhanced insulin secretion might be mediated through the IL6-mediated PLC-IP3 pathway as previously reported.15 Hence, inhibitors such as U73122 (PLC inhibitor), Neomycin (PIP2 inhibitor), and Xestospongin C (IP3 inhibitor) were used to evaluate the role of PLC-IP3 pathway in the PSC-cocultured Min6 cell function. Surprisingly, neither the GSIS nor the total insulin contents were altered in PSC-cocultured Min6 cells with inhibitors. The observations made from these studies suggest that the augmented insulin secretion from Min6 cells is driven by a mechanism, independent of IL6 secreted by activated PSCs in the coculture system. Apart from IL6, PSCs are also known to secrete factors such as activin-A, HGF, and endothelin1 in addition to acetylcholine, a neurotransmitter, which is known to promote the insulin secretion from the β-cells. However, the influence of these factors on Min6 cell insulin secretory function in the coculture system was not examined, which stands as a limitation of the present study.
We have studied the effect of PSCs on β-cell function with a background to understand the role of in vitro-activated PSCs on β-cell function in pancreatogenic diabetes associated with exocrine disease (CP), a pancreatic disease in which PSCs are activated and responsible for progressive fibrosis. The role of quiescent and activated PSCs on islet function is still a question of debate, both in physiological and in pathological condition. However, the presence of vitamin A and its metabolites (which are known to promote the insulin secretion from beta cells) in the lipid droplets of quiescent PSCs suggest the possible involvement of these cells in maintaining the β-cell mass and their function under physiological conditions.48, 49 More important, the recently identified quiescent islet stellate cell-secreted Wnt5a has been reported to improve the islet insulin secretion50 upon high glucose challenge, presenting the quiescent PSC as an “ally” in islet β-cell function. In contrast, the role of in vivo-activated PSCs on β-cell function, in addition to the initiation and progression of diabetes, is still uncertain. In view of the known ability of activated PSC factors that would promote as well as hinder the β-cell function, it is early to assess the role of activated PSCs in human disease based on this study. The dearth of knowledge of the role played by in vivo-activated PSCs on β-cell function in disease conditions warrant further investigations.
In conclusion, our findings suggest that the in vitro-activated PSCs potentiate the GSIS response from Min6 cells without altering the expression of β-cell-specific genes and miRNA-375. The augmented insulin secretion from Min6 cells is independent of IL6-mediated PLC-IP3-dependent pathway in the coculture system. Future studies directed toward understanding the glucose-dependent/independent effects of PSC secretions on β-cell insulin secretory function and assessing the activated PSC secretome may yield valuable information.
ACKNOWLEDGMENTS
Ratnakar Reddy Bynigeri is thankful to Department of Science and Technology for providing DST INSPIRE Fellowship (IF120436). We sincerely thank Professor Subramanyam Chivukula for English language review of the manuscript. We are thankful to Dr. Ravikanth VV and Mrs. Aparna J for helping with qPCR experiments. Also, we are grateful to Dr. Pavan Kumar P and Dr. Balkumar Reddy P for their help with IF microscopy and Flow Cytometry. We also thank Mr. Murali Manohar K for helping with the statistical analysis.
CONFLICT OF INTERESTS
All authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
RRB designed and conducted the experiments, analyzed the data, and wrote the manuscript. SM designed the experiments, analyzed the experimental data, corrected and approved the manuscript. RT, SSS, and NRD critically reviewed the manuscript and gave the intellectual inputs.