miR-496 remedies hypoxia reoxygenation–induced H9c2 cardiomyocyte apoptosis via Hook3-targeted PI3k/Akt/mTOR signaling pathway activation
Abstract
The hypoxia-reoxygenation (H/R) model helps analyze myocardial infarction triggered by acute myocardial ischemia, which induces cardiomyocyte proliferation and apoptosis. The Gene Expression Omnibus database was used to obtain the GSE74205 and GSE3866 microarray data, including microRNA (miRNA) and messenger RNA profiles, to catalog potential key miRNAs and genes. The role of rno-mir-496 expression in cardiomyocyte proliferation within 10 days of birth was established. The microRNA Target Prediction Database (miRDB) database—via Gene Ontology annotation—predicted hook microtubule tethering protein 3 (Hook3), a key target gene of rno-mir-496, was closely related to cell proliferation. Upregulation of miR-496 related to a significant reduction in apoptosis of H9c2 and human cardiomyocytes treatment with H/R. Moreover, transfection of H9c2 cells with miR-496 mimics, which were pretreated with H/R for 12 hours, increased Ki67 levels, proliferating cell nuclear antigen and Bcl-2 proteins; and decreased cleaved caspase-3 and Bax protein levels, as determined by reverse transcription-polymerase chain reaction and Western blot assays. A dual-luciferase reporter system confirmed that miR-496 targets the Hook3 suppressor. Hook3 overexpression stimulated apoptosis in H/R-treated cells, thus reducing cell proliferation. Upregulated miR-496 activated phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) signaling, while Hook3 exhibited the inverse trend in H/R-treated H9c2 cells. In summary, with Hook3 functionality's aid, miR-496 upregulation defends cells from H/R-induced apoptosis and stimulates cell proliferation. miR-496 targets Hook3 to trigger the PI3K/Akt/mTOR signaling pathway for antiapoptotic and proliferative effects.
Abbreviations
-
- AMI
-
- acute myocardial ischemia
-
- GEO
-
- Gene Expression Omnibus
-
- GO
-
- Gene Ontology
-
- H/R
-
- hypoxia-reoxygenation
-
- HCM
-
- human cardiomyocyte
-
- Hook3
-
- hook microtubule tethering protein 3
-
- I/R
-
- ischemia reperfusion
-
- IRI
-
- ischemia reperfusion–injury
-
- MI
-
- myocardial infarction
-
- MIRI
-
- myocardial ischemia reperfusion–injury
-
- miRNA
-
- microRNA
-
- PCNA
-
- proliferating cell nuclear antigen
1 INTRODUCTION
Acute myocardial ischemia (AMI) is a major cause of death in humans, via interruption of blood flow in the coronary artery.1 Myocardial ischemia reperfusion–injury (MIRI) in patients with AMI leads to cardiomyocyte apoptosis, myocardial necrosis, and death and thus, is the primary cause of unsatisfactory surgical results.2 A cardiomyocyte is a highly differentiated terminal cell, incapable of repairing myocardial injury via cell regeneration. In fact, cardiomyocyte apoptosis could expand the myocardial infarction (MI) range to cause ventricular remodeling.3, 4 Researchers and clinicians acknowledge this vicious cycle of cardiomyocyte apoptosis and ventricular remodeling, and thus emphasize on improving cardiomyocyte function to reduce MIRI-induced cardiomyocyte apoptosis, for better prognosis in patients with AMI.3 It is therefore imperative to identify the mechanisms of ischemia-reperfusion (I/R) induced cardiomyocyte apoptosis.
Previous research on MIRI indicates a principal role of microRNAs (miRNAs), the noncoding RNAs that significantly regulate cell differentiation, cycle, proliferation, and apoptosis, and are extensively expressed in different organs and tissues.5, 6 The miRNA biogenesis pathway is closely connected with heart functions and is involved in regulating angiogenesis,7 heart development,8 heart failure,9 and ischemia reperfusion–injury (IRI).7 miRNA expressions are considerably regulated in the myocardial ischemia process and hypoxia. For example, miR-99a has a positive effect, as it improves heart function in patients with AMI.10 According to the GSE74205 data analysis, miR-496 is significantly upregulated in rat cardiomyocytes, within 10 days of birth. A mammalian target of rapamycin (mTOR) regulator target correlated with aging, miR-496 is also expressed in acute myeloid leukemia, pancreatic adenocarcinoma, oropharyngeal cancer, and cerebral I/R injury. There has been no research, till date, on miR-496 function and mechanism of action in MIRI, thus leading us to investigate it in the context of IRI-induced cardiomyocyte apoptosis.
The human hook microtubule tethering proteins (Hooks) family comprises three homologs, (Hook1, Hook2, and Hook3, which were previously reported as organelle–microtubule linkers trafficking endocytic protein in many diseases.11-15 Hooks, with their separate subcellular localizations, naturally exhibit distinct cellular functionality. For example, Hook3 is closely related to poor prostate cancer prognosis,13 and regulates the timing of neurogenesis.15 Previous analysis has established Hook3 as the target gene of miR-496. This structured study on MI deploys the hypoxia-reoxygenation (H/R) model to explore the function of miR-496 and Hook3 in H9c2 cells and investigates whether miR-496 directly targets Hook3 to influence H/R-induced cardiomyocyte apoptosis.
2 MATERIALS AND METHODS
2.1 Bioinformatics analysis
Relevant microarrays were extracted from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), GSE74205 and GSE3866 to include miRNA and messenger RNA (mRNA) profiles. The authors consulted the miRDB (http://www.mirdb.org/) to predict miR-496 target genes, which included Hook3 and further ascertained the biological functionality potentially specific to miR-496-targeted genes via Gene Ontology (GO) (http://www.geneontology.org/) analysis.
2.2 Cell culture
The H9c2 rat cardiomyocyte cells were procured from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) and human cardiomyocyte (HCM) were purchased from the American Type Culture Collection (Manassas, VA). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone, Thermo Fisher Scientific, Inc, Wilmington, DE) augmented with 10% fetal bovine serum (FBS; HyClone; Thermo Fisher Scientific, Inc), at 37°C in a 5% CO2 incubator, maintaining the requisite humidity. When the cultivated cells attained 70% to 80% confluence, they were synchronized in serum-free DMEM for 12 hours, before the experiment.
2.2.1 Establishment of hypoxia-reoxygenation cell model and cell treatment
H9c2 and HCM were maintained in DMEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. To induce H/R injury, cells were incubated in a regulated hypoxic plastic chamber filled with a mixture of 95% N2 and 5% CO2 and replaced the media with fresh 1.5 g/L glucose DMEM without serum for 3 or 6 hours at 37°C, followed by reoxygenation under normoxic conditions (95% air and 5% CO2) at 37°C for 6 hours as previously described.16
Following is the cell treatment: H9c2 cells or HCM were treated with H/R for 9 hours (3 hours hypoxia and then 6 hours reoxygenation); H9c2 cells or HCM were treated with H/R for 12 hours (6 hours hypoxia and then 6 hours reoxygenation); H9c2 cells were transfected with miR-496 mimics/inhibitors and negative controls; H9c2 cells were transfected with Hook3 overexpression or siHook3 and negative controls; cotransfection of H9c2 cells with Hook3 overexpression plasmids and miR-496 mimics were treated with H/R for 9 hours (3 hours hypoxia and then 6 hours reoxygenation).
2.3 Transfection of miR-496 mimics and inhibitors
miR-496 mimics, inhibitors, and negative control oligonucleotides were chemically integrated (Shanghai GenePharma Co, Ltd, Shanghai, China), with Rattus norvegicus sapiens (rno)-miR-496 mimics: sense, 5′-GGUUAGAGGAAAGGUAGC-3′ and antisense, 5′-UCACAAGCUUACCUC-3′; and 5′-TGGTGTGTTCATTTTAT-3′, for the rno-miR-496 inhibitor. 6-Carboxyfluorescein-aminohexyl phosphoramidite–labeled small interfering RNA (siRNA) was used as a negative control. For transfection, 4 × 106 H9c2 cells were seeded per well in six-well plates and cultured to a confluence greater than 70%, in antibiotic-free medium for 48 hours. Cells were transfected under serum-free conditions for 6 hours with miR-496 mimics and inhibitor (40 nm) using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc) before the culture supernatant was entirely substituted with the complete medium.
2.4 Transient transfection of plasmid DNA or siRNA
OBIO Technology (Shanghai, China) designed and assembled the Hook3 overexpression and control plasmids (mock). siRNAs specific to Hook3 (Dharmacon) was used for a knockdown. Sequences for Hook3 siRNA were 5′-GGAGGAGUCUCUUCAGCAU-3′. Following were the sequences for Hook3 overexpression: sense, 5′-CGGGAGCGGCCGGACG-3′, antisense, 5′-AACTCCAATATAATTGAACTTAGAAT-3′. Using Lipofectamine 3000 (Invitrogen), H9c2 cells were transfected with Hook3 overexpression or siRNA-Hook3.
2.5 Cell counting kit-8 assay
After H9c2 cells and HCM were treated, the cell counting kit-8 (CCK-8) was used to gauge the H9c2 (a-g groups) cell viability via a convenient assay (C0038; Beyotime Institute of Biotechnology), wherein each well was supplemented with the CCK-8 reagent and incubated for 4 hours. A microplate reader set at 490 nm logged the absorbance. Experiments were performed in triplicates.
2.6 Lactate dehydrogenase assay
Cells were seeded in 24-well plates at a density of 2 × 105 cells/well and incubated with H/R-treated cells serum, at 37°C for 24 hours. The medium was then replaced with fresh Creek's minimal medium and reincubated at 37°C for 6 hours. A total of 120 µL of the supernatant was centrifuged at 12 000g and 4°C for 10 minutes and utilized for the lactate dehydrogenase (LDH) assay, as per the manufacturer's protocol (cat# C0016; Beyotime Institute of Biotechnology). Absorbance was measured at 490 nm with a plate reader (SpectraMax; Molecular Devices, LLC, Sunnyvale, CA).
2.7 Flow cytometry assay
Cells were washed three times with phosphate-buffered saline 48 hours after transfection and then resuspended for 15 minutes at room temperature with 5 µL each of annexin V-fluorescein isothiocyanate and propidium iodide (BD Biosciences, Franklin Lakes, NJ). The apoptotic population was measured with a CyAn ADP cytometer (Dako; Agilent Technologies, Inc, Santa Clara, CA). Accumulation of double-positive cells in the upper-right quadrant is related to apoptosis.
2.8 Dual-luciferase reporter gene assay
The miRNA bioinformatics tool miRDB helped predict the miR-496-Hook3 3′-untranslated region (3′-UTR) binding site. The plasmid extraction kit (Promega) instruction manual facilitated the fabrication of Hook3 3′-UTR wild-type (wt) and Hook3 3′-UTR mutant (mut)-type plasmids. H9c2 transfection via the Lipofectamine 3000 kit (Invitrogen) helped designate the respective cell groups: miR-496 mimics+Hook3-wt, miR-496 mimics+Hook3-mut, miR-496 negative control (NC)+PHook3-wt and miR-496 NC+Hook3-mut. The dual-luciferase reporter gene assay (Promega, Madison, WI) uses luciferase activity to subsequently present it as the relative Firefly/Renilla luciferase activity ratio.
2.9 Protein extraction and Western blot analysis
The total concentration of cell proteins extracted via the radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) was measured with the bicinchoninic acid assay (Beijing Solarbio Science & Technology Co, Ltd). A total of 30 µg protein samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% Tris-glycine), and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA) (Bio-Rad Laboratories, Inc). The membranes were blocked with 5% nonfat dry milk in Tris-HCl buffer saline (pH 7.4) containing 0.1% Tween-20 for 1 hour at room temperature. These membranes were incubated overnight at 4°C with primary antibodies against Hook3 (cat# K17936-UBK; Biomart Science and Technology Ltd; Beijing, China), Ki67 (cat# ab15580; Abcam), proliferating cell nuclear antigen (PCNA) (cat# ab92552; Abcam), cleaved caspase-3 (cat# ab49822; Abcam), Bax (cat# ab32503; Abcam), Bcl-2 (cat# ab59348; Abcam), phosphatidylinositol-3-kinase (PI3K) (cat# bs6423R; Bioss), protein kinase B (Akt) (cat# bsm-33325M; Bioss), mTOR (cat# ba1992R; Bioss), phospho-PI3K (cat# bs6417R; Bioss), phospho-Akt (cat# bs5194R; Bioss), and phospho-mTOR (cat# bs3495R; Bioss), and subsequently for 2 hours at room temperature with horseradish peroxidase-conjugated secondary antibodies (cat# ab6278; Abcam). Protein bands of interest were detected using a Super Signal Enhanced Chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc), and band quantification were performed via the densitometry and the ImageJ Pro 6.0 analyses systems (National Institutes of Health).
2.10 RNA extraction and reverse transcription-polymerase chain reaction
TRIzol reagent (Invitrogen) was used to isolated cell RNA and a PrimeScript reagent kit with genomic DNA Eraser (Takara) was used to reverse transcribe the complementary DNA (cDNA). Quantitative reverse transcription-polymerase chain reaction (RT-qPCR) was performed on an iCyler iQ Real-Time PCR System (Bio-Rad Laboratories Inc), using an SYBR Green PCR kit (TaKaRa). The relevant endogenous control genes U6 and glyceraldehyde 3-phosphate dehydrogenase helped identify miR-496, Hook3, Ki67, PCNA, Bax, and Bcl-2 mRNA. The miRNeasy Mini kit (Qiagen) was used to isolate total RNA for quantitative miRNA analysis, and the Bulge-Loop miRNA qRT-PCR Starter kit (Ribibio) was used to reverse-transcribe the mature miRNAs. All assays were conducted with strict adherence to the respective instruction manuals, and the reaction conditions set were 94°C for 2 minutes and again for 40°C for 10 seconds cycles, then 60°C for 1 minute, and the last being 72°C for 30 seconds. Table 1 presents the RT-qPCR primers. The method was used to compute the fold change.
| Gene | Primer sequence | Species |
|---|---|---|
| miR-496 | Forward: 5′-TCACAAGCTTACCTTTAACAA-3′ | Rat |
| Reverse: 5′-TTAGAGGAAATTCCGAATTTC-3′ | ||
| Hook3 | Forward: 5′-CATGACAGCCATCCAAGAGC-3′ | Rat |
| Reverse: 5′-CCTTCGCTGACAAAGCTTCA-3′ | ||
| Ki67 | Forward: 5′-GGACTCGCAGTTTGAGAAGG-3′ | Rat |
| Reverse: 5′-TGCAAATGTCCTCGTTTCTG-3′ | ||
| PCNA | Forward: 5′-GGATTCGTCTCACGTCTCCT-3′ | Rat |
| Reverse: 5′-TTGGACATGCTGGTGAGGTT-3′ | ||
| Bax | Forward: 5′-ACTAAAGTGCCCGAGCTGAT-3′ | Rat |
| Reverse: 5′-ATGGTGAGTGAGGCAGTGAG-3′ | ||
| Bcl-2 | Forward: 5′-GGCATCTTCTCCTTCCAGC-3′ | Rat |
| Reverse: 5′-TCCCAGCCTCCGTTATCC-3′ | ||
| U6 | Forward: 5′-GCTGGTGGAGCTGAAGAATG-3′ | Rat |
| Reverse: 5′-GCAGGTACTTGATGGTGCTG-3′ | ||
| GAPDH | Forward: 5′-AAGATGGTGAAGGTCGGTGT-3′ | Rat |
| Reverse: 5′-GCTTCCCATTCTCAGCCTTG-3′ |
- Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Hook, hook microtubule tethering protein; miR, microRNA; PCNA, proliferating cell nuclear antigen; PCR, polymerase chain reaction.
2.11 Statistical analysis
The statistical analysis was implemented via the GraphPad Prism 5.0 software (GraphPad Software, Inc, La Jolla, CA), all the experimental data are represented as mean ± standard deviation. The one-way variance analysis with the Tukey range test indicated P < .05 as statistically significant.
3 RESULTS
3.1 miR-496 was lowly expressed in H/R-treated H9c2 cells
To assess the validity of GSE74205 microarray data, 46 miRNAs in rat cardiomyocytes, within 10 days of birth, were selected. The selection included 45 significantly dysregulated miRNAs: upregulated (such as miR-496, miR-466b, miR-31, miR-23a, miR-146a, and miR-465) or downregulated (such as miR-494, let-7f, miR-542, miR-532-5p, miR-455, and miR-322), which were selected as possible candidates for further investigation (Figure 1A and 1B). The morphology of H9c2 cells treated with H/R (3/6 hours) (3 hours of hypoxia followed by 6 hours of reoxygenation) or H/R (6/6 hours) (6 hours of hypoxia followed by 6 hours of reoxygenation) was observed through an inverted microscope. The untreated H9c2 cells grew normally (Figure 1C). The LDH level significantly increased after H9c2 cells were treated with H/R (3/6 hours) or H/R (6/6 hours) (Figure 1D). The viability of H9c2 cells treated with H/R (6/6 hours) was obviously better than that of H9c2 cells treated with H/R (3/6 hours), meanwhile, the apoptosis level of H9c2 cells treated with H/R (6/6 hours) was significantly higher than that of H9c2 cells treated with H/R (3/6 hours) (Figure 1E-G). We found that miR-496 is poorly expressed in H9c2 cells treated with H/R (3/ 6 hours) or H/R (6/6 hours) (Figure 1H).

miR-496 was lowly expressed in H/R-treated H9c2 cells and HCM. A, Heatmap of the 39 differentially expressed miRNAs (38 upregulated and seven downregulated genes) from GSE74205 data. Green ones represent upregulation and red represent downregulation. B, Volcano plot of 46 genes. Red plots represented genes with fold change ≥1.5 or ≤ −1.5, P < .01. Black plots represent the rest of the genes with no significant expression change. C, The morphology of H9c2 cells treated with H/R (3/6 hours) (3 hours of hypoxia followed by 6 hours of reoxygenation) or H/R (6/6 hours) (6 hours of hypoxia followed by 6 hours of reoxygenation) was observed through an inverted microscope. D, The LDH level was detected after H9c2 cells were treated with H/R (3/6 hours) or H/R (6/6 hours). E, CCK-8 assay was used to detected cell viability in H9c2 cells were treated with H/R (3/6 hours) or H/R (6/6 hours). F,G, Flow cytometry assay was used to detect cell apoptosis in H9c2 cells were treated with H/R (3/6 hours) or H/R (6/6 hours). H, miR-496 mRNA level was detected by RT-PCR assay after H9c2 cells were treated with H/R (3/6 hours) or H/R (6/6 hours). I, The morphology of HCM treated with H/R (3/6 hours) (3 hours of hypoxia followed by 6 hours of reoxygenation) or H/R (6/6 hours) (6 hours of hypoxia followed by 6 hours of reoxygenation) was observed through an inverted microscope. J, The LDH level was detected after HCM were treated with H/R (3/6 hours) or H/R (6/6 hours). K, CCK-8 assay was used to detected cell viability in HCM were treated with H/R (3/6 hours) or H/R (6/6 hours). L,M, Flow cytometry assay was used to detect cell apoptosis in HCM and were treated with H/R (3/6 hours) or H/R (6/6 hours). N, miR-496 mRNA level was detected by RT-PCR assay after HCM were treated with H/R (3/6 hours) or H/R (6/6 hours). U6 was used as load control. Data are presented as the mean ± standard deviation. *P < .05 vs control cells group, #P < .05 vs H/R (3/6 hours ) cell group. CCK-8, cell counting kit-8; FITC, fluorescein isothiocyanate; HCM, human cardiomyocytes; H/R, hypoxia-reoxygenation; LDH, lactate dehydrogenase; miRNA, microRNA; mRNA, messenger RNA; RT-PCR, reverse transcription-polymerase chain reaction
3.2 miR-496 was lowly expressed in H/R-treated HCM
Similarly, the morphology of HCM treated with H/R (3/6 hours) (3 hours of hypoxia followed by 6 hours of reoxygenation) or H/R (6/6 hours) (6 hours of hypoxia followed by 6 hours of reoxygenation) were observed through an inverted microscope. The untreated HCM grew normally (Figure 1I). The LDH level significantly increased after HCM were treated with H/R (3/6 hours) or H/R (6/6 hours) (Figure 1G). The viability of HCM treated with H/R (6/6 hours) was obviously better than that of HCM treated with H/R (3/6 hours), meanwhile, the apoptosis level of HCM treated with H/R (6/6 hours) was significantly higher than that of HCM treated with H/R (3/6 hours) (Figure 1K-M). We found that miR-496 is poorly expressed in HCM treated with H/R (3/6 hours) or H/R (6/6 hours) (Figure 1N).
3.3 Overexpressed miR-496 reduced H/R-induced apoptosis in H9c2 cells and regulated the apoptosis and proliferation-related gene expressions
miR-496 was significantly amplified in the miR-496 mimic group, and diminished in miR-496 inhibitor group when compared with Blank and Blank NC groups (Figure 2A). Upregulated miR-496 reduced the H/R-induced apoptosis, and fostered cell proliferation, whereas downregulation of miR-496 increased cellular apoptosis and hindered proliferation in H/R-treated H9c2 cells (Figure 2B-D). The expression levels of the genes, Ki67 and PCNA, associated with proliferation, significantly dwindled with H/R, whereas miR-496 mimics increased protein expression significantly (Figure 2E). The expression levels of the apoptosis-inducing cleaved caspase-3, Bax, and Bcl-2 genes were highly H/R-regulated; miR-496 mimics notably diminished the protein expressions of the cleaved caspase-3 and Bax genes while increasing Bcl-2 protein expression (Figure 2E). Both mRNA and protein expression levels of the genes exhibited a correspondingly similar tendency (Figure 2F).
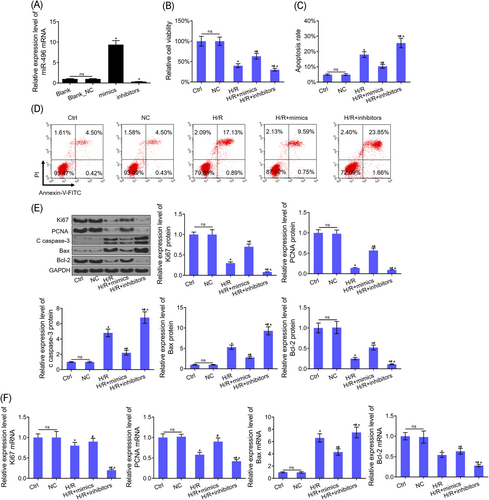
miR-496 regulated the viability and apoptosis in H9c2 cells. A, The expression level of miR-496 mRNA was detected by RT-PCR after H9c2 cells treated with mimics/inhibitors/NC. B, CCK-8 assay was used to detected cell viability in the cells of every group. C, D, Flow cytometry assay was used to detected cell apoptosis in the cells of every group. E, The protein levels of Ki67, PCNA, cleaved caspase-3, Bax, and Bcl-2 were detected by Western blot assay in the cells of every group. F, The mRNA levels of Ki67, PCNA, Bax, and Bcl-2 were detected by RT-PCR assay in the cells of every group. U6 and GAPDH were used as load control, respectively. Data are presented as the mean ± standard deviation. *P < .05 vs ctrl or NC group, #P < 0.05 vs H/R group and ^P < .05 vs H/R+mimics group. CCK-8, cell counting kit-8; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; miR, microRNA; mRNA, messenger RNA; NC, negative control; PCNA, proliferating cell nuclear antigen; RT-PCR, reverse transcription-polymerase chain reaction
3.4 Hook3 was a target gene of miR-496
The analysis of DNA microarray from the GSE3866 database, in which a total of 839 differentially expressed genes (DEGs) were verified in rat heart tissues (Figure 3A and 3B). GO analysis showed that 839 DEGs mainly enrich in “negative regulation of the cellular process,” “cell development,” “cell proliferation,” “system development,” and “biological regulation” (Figure 3C). A total of 164 genes were considered to be the target genes of miR-496 by using bioinformatics tool miRDB database (http://www.mirdb.org/) (Table 2). Furthermore, APAF1 was both a DEG from GSE3866 database and a target gene of miR-496 from miRDB database (Figure 3D). Mechanically, the binding sites and modified sequence in the APAF1 3′-UTR are shown in Figure 3E. Dual-luciferase reporter assay suggested that the luciferase activity of H9c2 cells with APAF1-WT transfection was significantly decreased by miR-496 mimics and increased by miR-496 inhibitors (Figure 3F). In addition, miR-496 overexpression inhibited Hook3 mRNA and protein levels, whereas, miR-496 inhibition upregulated Hook3 mRNA and protein levels in H9c2 cells (Figure 3G and 3H).
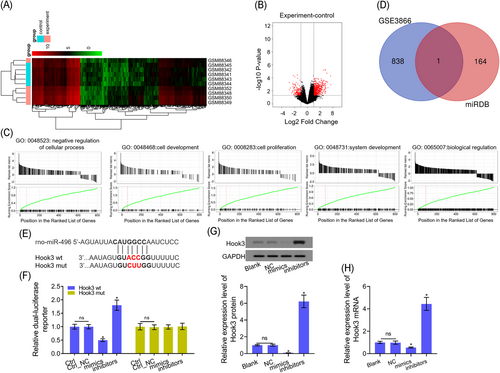
Hook3 is a target gene of miR-496. A, Heatmap of the 839 DEGs (653 upregulated and 186 downregulated genes) from GSE3866. Green ones represented upregulation and red represented downregulation. B, Volcano plot of 839 genes. Red plots represent genes with fold change ≥1.5 or ≤ −1.5, P < .01. Black plots represent the rest of the genes with no significant expression change. C, GO annotations analysis revealed that 839 DEGs were implicated in a broad range of biological processes, cell component and molecular function. D, Venn diagram of one overlapping 839 DEGs target and 164 target genes in the two databases. E, The miR-496 binding sequence in the 3′-UTR of Hook3. F, The luciferase reporter assay was performed to detect the luciferase activities of the Hook3 3′-UTR in H9c2 cells. G,H, Hook3, mRNA, and protein levels were detected by Western blot assay and RT-PCR after H9c2 cells were transfected with miR-496 mimics/inhibitors. GAPDH was used as load control, respectively. Data are presented as the mean ± standard deviation. *P < .05 vs blank, ctrl or NC group. 3′-UTR, 3′-untranslated region; DEG, differentially expressed gene; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GO, Gene Ontology; Hook, hook microtubule tethering protein; miR, microRNA; mRDB, XXX; mRNA, messenger RNA; mut, mutant; RT-PCR, reverse transcription-polymerase chain reaction; wt, wild-type
| No. | Gene name | No. | Gene name |
|---|---|---|---|
| 1 | Foxo1 | 83 | Taf5 |
| 2 | Sec. 14l1 | 84 | Il1a |
| 3 | Daam2 | 85 | Rsf1 |
| 4 | Ptpn12 | 86 | Plod1 |
| 5 | Amfr | 87 | Sntb1 |
| 6 | Hivep2 | 88 | Hspd1 |
| 7 | Spag6l | 89 | Glra2 |
| 8 | Epb41l2 | 90 | Lclat1 |
| 9 | Slc34a2 | 91 | Atad2 |
| 10 | Rock2 | 92 | Dpp3 |
| 11 | Slc1a2 | 93 | Chp1 |
| 12 | Arid4b | 94 | Tmem115 |
| 13 | Fam175a | 95 | LOC678760 |
| 14 | Eif5a2 | 96 | Setd7 |
| 15 | Tfap2b | 97 | Tspan11 |
| 16 | Reck | 98 | Fam26e |
| 17 | Degs2 | 99 | Ccl9 |
| 18 | Fam9b | 100 | Plp1 |
| 19 | LOC100912163 | 101 | Itga6 |
| 20 | Noa1 | 102 | LOC102546492 |
| 21 | Ccdc67 | 103 | Otub2 |
| 22 | Dhdh | 104 | Dcdc2 |
| 23 | Trdn | 105 | Rqcd1 |
| 24 | Rfx7 | 106 | Lrrc20 |
| 25 | Nedd1 | 107 | Ssbp2 |
| 26 | Lipf | 108 | Fam103a1 |
| 27 | Crispld1 | 109 | Npy1r |
| 28 | Eif4g2 | 110 | Adcyap1 |
| 29 | Eif3j | 111 | Rhbdf1 |
| 30 | Rps6kb1 | 112 | Btaf1 |
| 31 | Lypla1 | 113 | Morc3 |
| 32 | Sepsecs | 114 | RGD1560672 |
| 33 | Caps2 | 115 | Pgrmc2 |
| 34 | LOC102548067 | 116 | Gpihbp1 |
| 35 | Hook3 | 117 | LOC691414 |
| 36 | Chek1 | 118 | Nmnat1 |
| 37 | LOC100360205 | 119 | Setd5 |
| 38 | Mdm2 | 120 | Rbm20 |
| 39 | Pno1 | 121 | Pigzl1 |
| 40 | Ankfn1 | 122 | LOC501396 |
| 41 | Naaladl2 | 123 | Arl13b |
| 42 | Kcne2 | 124 | Fam83h |
| 43 | B3galt2 | 125 | Syngr3 |
| 44 | LOC102550296 | 126 | Pcyt1b |
| 45 | Gabpb2 | 127 | Clca5 |
| 46 | Lipk | 128 | Slc17a3 |
| 47 | Trim23 | 129 | LOC100360457 |
| 48 | LOC100910771 | 130 | Yipf6 |
| 49 | Sh3d19 | 131 | LOC100909505 |
| 50 | Capza1 | 132 | Gpr126 |
| 51 | Adam12 | 133 | Gars |
| 52 | Impa1 | 134 | Pax8 |
| 53 | Zfp189 | 135 | LOC102556552 |
| 54 | Wee2 | 136 | Fam196b |
| 55 | Map3k7 | 137 | FAM120C |
| 56 | Dsg1 | 138 | LOC691195 |
| 57 | Atxn7 | 139 | C8b |
| 58 | Sostdc1 | 140 | Nfkbib |
| 59 | LOC690826 | 141 | Alx3 |
| 60 | Gtl3 | 142 | Ric8a |
| 61 | Tmem126a | 143 | Smarcd1 |
| 62 | Mog | 144 | Kti12 |
| 63 | Ppp1r8 | 145 | Siglech |
| 64 | Plcxd2 | 146 | Dnajc6 |
| 65 | Rab3a | 147 | Slc9a1 |
| 66 | Pgm2l1 | 148 | Tra2a |
| 67 | LOC102556832 | 149 | LOC102552823 |
| 68 | Naa15 | 150 | RGD1310352 |
| 69 | Lmo3 | 151 | Vom2r75 |
| 70 | Rnf151 | 152 | Pign |
| 71 | Erlin2 | 153 | Tbc1d9 |
| 72 | Tbx15 | 154 | Kcnd2 |
| 73 | Wasf3 | 155 | Grid2 |
| 74 | Casp8ap2 | 156 | F3 |
| 75 | RGD1310335 | 157 | LOC102552190 |
| 76 | Klf3 | 158 | LOC690911 |
| 77 | Maml2 | 159 | Tgfbrap1 |
| 78 | LOC100910646 | 160 | LOC102547413 |
| 79 | Rai2 | 161 | Uspl1 |
| 80 | LOC691352 | 162 | LOC100910007 |
| 81 | LOC500213 | 163 | Vom2r31 |
| 82 | Nhlh2 | 164 | Sel1l |
- Abbreviation: miRDB, XXX.
3.5 Hook3 regulated H/R-induced apoptosis in H9c2 cell
First, Hook3 protein and mRNA levels were significantly increased after H9c2 cells were transfected with Hook3 overexpression, meanwhile, Hook3 protein and mRNA levels were significantly decreased after H9c2 cells were transfected with siRNA-Hook3 (Figure 4A and 4B). Cell viability was inhibited by H/R treatment. However, the effect of H/R inhibiting cell viability was further aggravated by Hook3 overexpression and was reversed by Hook3 inhibition partly (Figure 4C and 4D). The flow cytometric analysis indicated that Hook3 overexpression further upregulated H/R-induced cell apoptosis, and Hook3 inhibition partly inhibited H/R-induced cell apoptosis (Figure 4E and 4F).
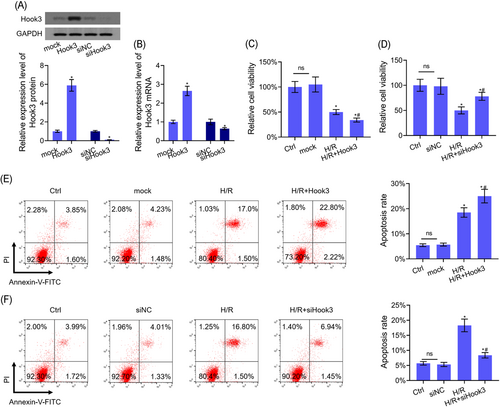
Hook3 regulated the viability and apoptosis in H9c2 cells. A,B, Hook3, mRNA, and protein levels were detected by Western blot and RT-PCR assays after H9c2 cells were transfected with Hook3 overexpression or inhibition. C,D, CCK-8 assay was used to detected cell viability after H9c2 cells were transfected with Hook3 overexpression or inhibition. E,F, Flow cytometry assay was used to detect the apoptosis level after H9c2 cells were transfeced with Hook3 overexpression or inhibition. GAPDH was used as load control. Data are presented as the mean ± standard deviation. *P < .05 vs mock or siNC group and #P < .05 vs H/R group. CCK-8, cell counting kit-8; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; FITC, fluorescein isothiocyanate; Hook, hook microtubule tethering protein; H/R, hypoxia-reoxygenation; mRNA, messenger RNA; NC, negative control; RT-PCR, reverse transcription-polymerase chain reaction
3.6 H/R-induced cell apoptosis was reduced when miR-496 directly targets Hook3 in H9c2 cells
Our results showed that there was a totally different tread in cell viability and apoptosis between Hook3 overexpression and miR-496 overexpression (Figure 5). The functions of Hook3 overexpression inhibiting cell viability and promoting cell apoptosis in H/R-treated cells were abolished by miR-496 overexpression, whereas, the functions of miR-496 overexpression promoting cell viability and inhibiting cell apoptosis in H/R-treated cells were reversed by Hook3 overexpression (Figure 5A and 5C). The functions of Hook3 inhibition promoting cell viability and inhibiting cell apoptosis in H/R-treated cells were abolished by miR-496 inhibition, whereas, the functions of miR-496 inhibition inhibiting cell viability and promoting cell apoptosis in H/R-treated cells were reversed by Hook3 inhibition (Figure 5B and 5D).
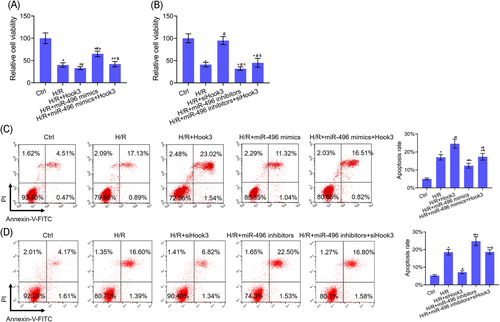
H/R-induced cell apoptosis reduced in H9c2 cells when miR-496 directly targeted Hook3. A, CCK-8 assay was used to detect cell viability after H9c2 cells were cotransfected Hook3 and miR-496 overexpressions. B, CCK-8 assay was used to detect cell viability after H9c2 cells were cotransfected siHook3 and miR-496 inhibitors. C, Flow cytometry assay was used to detect cell apoptosis after H9c2 cells were cotransfected Hook3 and miR-496 overexpressions. D, Flow cytometry assay was used to detected cell apoptosis after H9c2 cells were cotransfected siHook3 and miR-496 inhibitors. Data are presented as the mean ± standard deviation. *P < .05 vs ctrl group, #P < .05 vs H/R group, ^P < .05 vs H/R+Hook3/siHook3 group, and &P < .05 vs H/R+miR-496 mimics/inhibitors. CCK-8, cell counting kit-8; FITC, fluorescein isothiocyanate; Hook, hook microtubule tethering protein; H/R, hypoxia-reoxygenation; miR, microRNA
3.7 miR-496 targets Hook3 directly to regulate the expression levels of apoptosis- and proliferation-related genes, and activate PI3K/Akt/mTOR signaling pathway
Hook3 overexpression aggravated H/R-mediated inhibition of Ki67, PCNA, and Bcl-2 expression levels, and H/R-mediated increased of caspase-3 and Bax expression levels in H9c2 cells by Western blot and RT-PCR assays. Meanwhile, miR-496 overexpression promoted the Ki67, PCNA, and Bcl-2 expression levels, and inhibited caspase-3 and Bax expression levels in H/R-treated H9c2 cells by Western blot and RT-PCR assays. The functions of miR-496 or Hook3 regulating Ki67, PCNA, c caspase-3, Bax, and Bcl-2 expressions were reversed by cotransfection of miR-496 mimics and Hook3 overexpression in H9c2 cells (Figure 6A and 6B). Furthermore, Hook3 overexpression further inhibited H/R-induced inhibition of PI3K/Akt/mTOR signaling pathway. However, miR-496 overexpression could reverse H/R-induced inhibition of PI3K/Akt/mTOR signaling pathway. As expected, there was no significant difference in the activation of PI3K/Akt/mTOR signaling pathway between H/R-treated H9c2 cells and cotransfection of H9c2 cells with miR-496 mimics and Hook3 overexpression (Figure 6C).
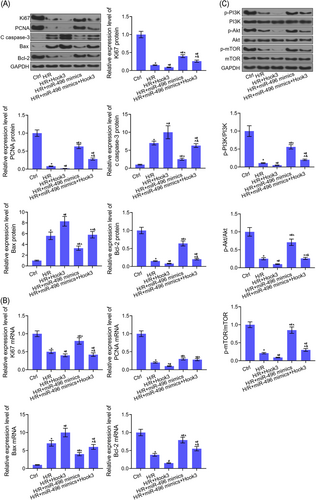
miR-496 directly targeted Hook3 to regulate the expression levels of apoptosis and proliferation-related genes and the activation of PI3K/Akt/mTOR signaling pathway. A, The protein levels of Ki67, PCNA, cleaved caspase-3, Bax, and Bcl-2 were detected by Western blot assay after H9c2 cells were cotransfected with Hook3 and miR-496 overexpressions. B, The mRNA levels of Ki67, PCNA, Bax, and Bcl-2 were detected by RT-PCR assay after H9c2 cells were cotransfected with Hook3 and miR-496 overexpressions. C, The phosphorylation level of PI3K, Akt, and mTOR, and the expression levels of PI3K, Akt, and mTOR proteins were detected by Western blot assay after H9c2 cells were cotransfected with Hook3 and miR-496 overexpressions. GAPDH was used as load control. Data are presented as the mean ± standard deviation. *P < .05 vs ctrl group, #P < .05 vs H/R group, ^P < .05 vs H/R+Hook3 group, and &P < .05 vs H/R+miR-496 mimics. Akt, protein kinase B; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Hook, hook microtubule tethering protein; H/R, hypoxia-reoxygenation; mTOR, mammalian target of rapamycin; miR, microRNA; mRNA, messenger RNA; PCNA, proliferating cell nuclear antigen; PI3K, phosphatidylinositol-3-kinase; RT-PCR, reverse transcription-polymerase chain reaction
4 DISCUSSION
miRNAs are essential for the development of the cardiovascular system and associated pathology and hold great potential as a means of treatment for cardiovascular disease.6, 8-10 Chen's17 study showed how miR-221 upregulation inhibits H/R-induced autophagy, and Wu's18 study revealed that miR-101 downregulation attenuates H/R-induced apoptosis in heart cells. These research results suggest that abnormal miRNA expressions are critical factors that regulate cell death and apoptosis in ischemic heart disease. miRNAs aggravate or reduce MIRI by regulating related gene expression.19 These studies lend a fresh insight to the field of cardiovascular pathologies. GSE74205 data set was downloaded from the GEO database. miR-496 was verified to be a DE-miRNA related to the growth of cardiomyocyte of rats within 10 days of birth. H/R-treated H9c2 cells and HCM were used to imitate MI-mediated MIRI in vitro. We need to further verify whether miR-496 affects H/R-induced apoptosis in heart cells. Our result showed that miR-496 was significantly downregulated in H/R-treated H9c2 cells and HCM (Figure 1). It showed that miR-496 should play the same role in rats and humans. In addition, we found that miR-496 overexpression could inhibit H/R-induced cell apoptosis, whereas, miR-496 inhibition could promote H/R-induced cell apoptosis (Figure 2A-D). It reported that miR-496 overexpression reduced cerebral I/R injury by inhibiting the expression level of Bcl-2 family proteins,20 in addition, proliferation indicators, such as PCNA and Ki67, decreased in I/R damage model,21 and activated caspase-3 promoted myocardial apoptosis in IRI.22, 23 Our results showed that miR-496 overexpression reduced the H/R-induced inhibition of PCNA, Ki67, and Bcl-2, and H/R-induced increased of caspase-3 and Bax. Therefore, miR-496 overexpression promoted cell viability and inhibited cell apoptosis via upregulating the proliferation and antiapoptosis protein expressions and downregulating proapoptosis protein expressions (Figure 2E and 2F). We conclude that upregulation of miR-496 relieved MRI in MI through inhibiting myocardial apoptosis. The regulation effect of miRNAs in cell proliferation and apoptosis is realized by binding target genes.5, 10, 18 Therefore, the target gene of miR-496 in H9c2 cells need to be verified by the miRDB database.
What is interesting about Hook3 is that it is only one overlapped gene from GSE3866 data set and miRDB database. Our results verified that miR-496 could target to bind Hook3 by RT-PCR, Western blot, and dual-luciferase activity assay (Figure 3). Furthermore, there were the opposite effects in H/R-induced cell apoptosis between Hook3 overexpression and miR-496 overexpression (Figure 4). This is an indication that Hook3 further aggravated MI-mediated MIRI by promoting myocardial apoptosis. It reported that Hook3, as a cargo adapter involved in Golgi and endosome transport, was mTOR regulator target.13, 15, 24 Based on the above research results, we needed to explore that miR0496 regulate H/R-induced cell apoptosis via binding to Hook3 to regulate mTOR signaling pathway. Our results showed that the effects of Hook3 and miR-496 in H/R-induced cell apoptosis were so opposed that they just cancel each other out (Figure 5). It showed that miR-496 inhibited H/R-induced cell apoptosis via targeting Hook3. Thus, miR-496 relieved MI-mediated MIRI via inhibiting the expression of Hook3.
mTOR is a downstream effector protein of the PI3K/Akt signaling pathway.25 Activated PI3K/Akt/mTOR signaling pathway is involved in cell proliferation, apoptosis and differentiation,22, 25 and could alleviate IRI in brain, renal, and heart.26-28 It was reported that miR-496 was considered to be a potentially relevant regulator of mTOR.29 Further study was needed to determine whether miR-496 regulating PI3K/Akt/mTOR signaling pathway via targeting Hook3. Our results showed that the functions of miR-496 overexpression increasing Ki67, PCNA, and Bcl-2 expressions, and decreasing caspase-3 and Bax expressions in H/R-treated H9c2 cells were reversed by Hook3 overexpression. These results showed that miR-496 regulated the expression levels of the proliferation- and apoptosis-related proteins by targeting Hook3 to play a role in the regulation of H/R-induced apoptosis. Furthermore, miR-496 overexpression reversed the functions of H/R treatment inhibiting the activation of PI3K/Akt/mTOR signaling pathway, and Hook3 overexpression promoted the functions of H/R treatment inhibiting the activation of PI3K/Akt/mTOR signaling pathway. After H/R-treated H9c2 cells were cotransfected with miR-496 mimics and Hook3 overexpression, the functions of the two genes in H9c2 cell treatment with H/R were counteracted each other to a certain extent (Figure 6). These results showed that miR-496 reduced H/R-induced cell apoptosis by binding to Hook3 to inhibit the activation of PI3K/Akt/mTOR signaling pathway. Therefore, miR-496 activated PI3K/Akt/mTOR signaling pathway via targeting inhibition of Hook3 to reduced MI-mediated IRI.
Apoptosis and autophagy regulate cell survival.30 It was reported that PI3K/Akt/mTOR signaling pathway is involved in autophagy.31 The above results had proved that miR-496 activated PI3K/Akt/mTOR signaling pathway via targeting the inhibition of Hook3 to reduce cell apoptosis in MI-mediated IRI. However, miR-496 regulating PI3K/Akt/mTOR signaling pathway in IRI-induced autophagy has not yet been studied. Therefore, we will explore the role of miR-496 in IRI-induced autophagy and related molecular mechanisms.
5 CONCLUSION
This study showed that H/R treatment inhibited miR-496 mRNA level and increased Hook3 protein and mRNA levels. miR-496 exercises a defensive function in H/R-treated myocardial cells by targeting Hook3 downregulation that suppresses cell apoptosis. Furthermore, miR-496 targets Hook3 to activate the PI3K/Akt/mTOR signaling pathway for antiapoptotic and proliferative effects.
ACKNOWLEDGMENTS
GSE74205 and GSE3866 data sets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) were used to perform the study. The project is supported with fund from Zhejiang Province Ministry of Education (No. Y20178744).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
YJ wrote the main manuscript and analyzed the data. YJ and SN performed the experiments. YJ and SN designed the study. All authors read and approved the final manuscript.
DATA ACCESSIBILITY
The data sets used and/or analyzed during the present study are available from the corresponding author on reasonable request.