Long noncoding RNA OIP5-AS1 aggravates cell proliferation, migration in gastric cancer by epigenetically silencing NLRP6 expression via binding EZH2
Abstract
The critical role of long noncoding RNAs (lncRNAs) in the development of multiple cancers has been revealed either functioning as a tumor initiator or a cancer suppressor. A widely recognized OIP5 antisense RNA 1 (lncRNA OIP5-AS1) has been validated to be an essential regulator of the tumorigenesis of various malignancies. Whereas, the potential role and the exact mechanism of lncRNA OIP5-AS1 by which OIP5-AS1 mediates gastric cancer (GC) progression remains vague. Therefore, first our work probed its expression levels in GC cell lines and related normal cells by real-time quantitative polymerase chain reaction. The heightened level of OIP5-AS1 was detected in GC cell lines. In terms of its cellular effects, we performed a series of functional experiments and as presented in the assays, the proliferative potential and motility was diminished. However, more apoptotic cells were induced with the introduction of OIP5-AS1 silencing. Meanwhile, higher Nod-like receptor pyrin domain-containing protein 6 (NLRP6) and enhancer of zeste homolog 2 (EZH2) expression in the GC cells was monitored. Besides, OIP5-AS1 was disclosed to locate mainly in the nucleus. In terms of mechanism, OIP5-AS1 directly bound to EZH2 and obstructed NLRP6 expression, speeding up GC progression.
1 INTRODUCTION
Gastric cancer (GC) is noted for being the most frequently occurring gastrointestinal tumor in East Asia and ranks as the major reason of cancer mortality globally.1, 2 What is worse, most patients are diagnosed as GC at an advanced stage, which unfortunately is accompanied by poor prognosis.3 Even though improvements in multiple fields have been made, little progress in early diagnosis and unsatisfactory treatment results of GC envelops people. Thus, screening and discovering novel and better biomarkers for GC early diagnosis and reliable management is imperative at present.
It has been depicted in multiple emerging research that long noncoding RNAs (lncRNAs), functional noncoding RNA molecules longer than 200 nucleotides, are of vital significance in the pathophysiological process of diverse human cancer progression, including GC. For example, lncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma.4 lncRNA MINCR activates Wnt/β-catenin signals to promote cell proliferation and migration in oral squamous cell carcinoma.5 lncRNA PICART1 inhibits cell proliferation by regulating the phosphoinositide-3-kinase/AKT and mitogen-activated protein kinase/extracellular-signal-regulated kinase signaling pathways in GC.6 Besides, as reported in accumulating data, lncRNAs managed to actuate a large amount of cancer phenotypes through multiple ways, which include microRNA (miRNA) sponging, epigenetic modification, RNA decay, and transcription regulation.7, 8 Developing cancer research discloses that epigenetic alterations containing histone modifications and DNA methylation constantly exist in tumor carcinogenesis and are widely investigated.9-11 Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase, whose upregulation was reportedly correlated with tumor initiation and discouraging clinical outcomes.12, 13 The EZH2-increased trimethylation of histone H3 lysine 27 (H3K27) on the promoters of target gene was also documented in previous studies.14, 15 And lncRNA-mediated epigenetic inhibition of antitumor gene via binding EZH2 in various cancers has been gradually detected. lncRNA-ANCR promotes osteosarcoma cell growth by interacting with EZH2 and repressing the expression of p21 and p27.16 TUG1 confers adriamycin resistance in acute myeloid leukemia by epigenetically suppressing miR-34a expression via EZH2.17 lncRNA AWPPH suppresses SMAD4 expression via EZH2 to accelerate bladder cancer progression.18
Recently, a newly identified lncRNA OIP5-AS1 has been characterized to promote the tumorigenesis of osteosarcoma,19 cervical cancer,20 lung cancer,21 colorectal cancer,22 and bladder cancer.23 No verification about its association or effect in GC has been included in preliminary works.
Therefore, the current study was designed to measure the expression level, detect specific function, and further certify the elusive molecular mechanism of lncRNA OIP5-AS1 in GC, which we expected to trigger new thoughts on GC diagnosis and therapy.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
GC cell lines (SGC7901, AGS, MKN45, and BGC803) purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China) and normal gastric epithelial GES-1 cells, acquired from the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences (Beijing, China) were involved in the study. Cultivation of these cell lines was performed in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Carlsbad, CA) incorporating 10% fetal bovine serum (FBS; HyClone, Logan, UT) in an atmosphere of 5% CO2 at 37℃.
2.2 Cell transfection
For the interference of OIP5-AS1, EZH2 and Nod-like receptor pyrin domain-containing protein 6 (NLRP6) expression, short hairpin RNAs specifically targeting OIP5-AS1, EZH2, and NLRP6 (sh-OIP5-AS1#1, sh-OIP5-AS1#2, sh-OIP5-AS1#3, sh-EZH2, sh-NLRP6) and matched negative control (sh-NC) were purchased from GenePharma (Shanghai, China). When the cell confluence reached 70%, transfection of all the plasmids and empty vector into GC cells was executed using Lipofectamine 2000 bought from Invitrogen, based on the manufacturer's instructions.
2.3 Real-time quantitative polymerase chain reaction
Based on the manufacturer's guide, total RNA extraction from GC cells was implemented by using TRIzol reagent purchased from Invitrogen. For the complementary DNA (cDNA) synthesis of lncRNA and messenger RNA (mRNA), the Reverse Transcription Kit (Takara, Otsu, Japan) was utilized. An SYBR Green PCR Kit (Takara) was applied for the quantification of lncRNA and mRNA. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels were used for the normalization of OIP5-AS1, EZH2, and NLRP6 expression. Primer sequences were as follows: OIP5-AS1 forward, 5′-TGCGAAGATGGCGGAGTAAG-3′ and reverse, 5′-TAGTTCCTCTCCTCTGGCCG-3′; NLRP6forward, 5′-CAGTACCTGGTGGGTATG-3′ and reverse, 5′-CCTGAAGCTCCTGTAGTG-3′; EZH2 forward, 5′-TGCACATCCTGACTTCTGTG-3′ and reverse, 5′-AAGGGCATTCACCAACTCC-3′; GAPDH forward, 5′-TGAACGGGAAGCTCACTGG-3′ and reverse, 5′-TCCACCACCCTGTTGCTGT-3′. Three times of real-time quantitative polymerase chain reaction (qRT-PCR) reactions were required.
2.4 Western blot analysis
Cells were harvested and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (RIPA; Beyotime, China) for this analysis. The protein from the lysates was obtained and its concentration was measured by bicinchoninic acid (BCA) kit (Beyotime, China). Afterward, an equal amount of protein samples was first separated via 8 or 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then separated proteins were transferred to polyvinylidene fluoride membranes (Millipore, Schwalbach, Germany). The protein after being blocked in 5% nonfat milk for 2 hours was incubated at 4℃ overnight with the following primary antibodies: anti-NLRP6 (Abcam), anti-EZH2 (Abcam), anti-GAPDH (Abcam). Then, the appropriate secondary was used for additional incubation with membranes. Eventually, Enhanced Chemiluminescence Kit (Pierce, Waltham, MA) was utilized for analyzing the blots signals of the assays repeated three times with Image Lab Software applied for related data analysis.
2.5 Cell counting kit-8 assay
The estimation of proliferation rate of AGS and SGC7901 cells was conducted by means of cell counting kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan). Ninety-six-well plates were seeded with GC cells (2 × 103 cells per well) followed by the addition of 10 μL of CCK-8 solution into each well every day at the specific time point. After 2 hours of incubation, the optical density at 450-nm wavelength was assessed by a Synergy H1 microplate reader (BioTek) every 24 hours for 5 days. Three independent experiments were carried out following the manufacturer's protocol.
2.6 Flow cytometry apoptosis analysis
Forty-eight hours after transfection, flow cytometry apoptosis analysis was conducted to evaluate GC cell apoptosis using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences) as instructed by the manufacturer's manuals. First, 1 × 106 GC cancer cells in each well were rinsed by 500 μL binding buffer and for the fixation of GC cells, precooled 70% ethanol was added. Next, 5 μL Annexin V-FITC and 5 μL PI containing RNAase was utilized for staining apoptotic cells without exposure to light. Lastly, under the excitation wavelength of 488 nm, the apoptosis status of GC cells was monitored by flow cytometry. Assays performed in triplicate were to obtain mean values.
2.7 Transwell assay
GC cell migration capacity was tested by transwell migration assay in a 24-well noncoated Matrigel transwell chamber (8-μm pore size; Corning, MA). In brief, the GC cells plated in the top chamber were cultured in serum-free media. The Lower chamber of a 24-well plate was filled with RPMI 1640 medium incorporating 20% FBS. After culturing for 24 hours to allow for migration, we removed the cells which remained on the upper surface of the membrane with a cotton swab and the migrating cells were immobilized by ethanol and stained with crystal violet. The migrating or invading cells were imaged and counted under a fluorescent microscope in five random microscopic fields.
2.8 RNA immunoprecipitation assay
As instructed by the manufacturer's protocol, the RIP experiments repeated three times in this section were carried out using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA). The AGS and SGC7901 cells, when they reached about 80% cell confluence, were bathed twice with phosphate-buffered saline (PBS) and trypsinized. After incubation with RIP lysis buffer and centrifugation, total cell extracts were incubated in RIP buffer with magnetic beads conjugated with Anti-EZH2 antibody (Abcam) at 4℃ for 18 hours to recruit RNA-protein complexes. Next, protein RNA complexes associated with anti-EZH2 antibody were rinsed three times in NT2 buffer and underwent DNase I treatment. qRT-PCR analysis was used for detecting coprecipitated RNAs. Total RNAs (input controls) and IgG (negative controls) were also detected for the confirmation of the signals that were caused by RNAs specifically binding to EZH2.
2.9 Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was carried out with the application of the SimpleChIP® Enzymatic Chromatin IP Kit (CST) and after formaldehyde treatment and incubation, DNA-protein crosslinks were generated. Antibodies including anti-EZH2 (Millipore), anti-H3K27me3 (Millipore), corresponding anti-IgG (abcam), and anti-DNA polymerase II (sigma) were also utilized in this assay with anti-IgG and anti-DNA polymerase II, respectively, serving as the negative control or positive control. GC cells were first lysed in RIPA lysis buffer. Crosslinked chromatin after sonication was sheared into 200-1000 bp fragments through enzymatic digestion. After centrifugation, chromatin was immunoprecipitated with the above recognizing antibodies overnight at 4℃. Then, protein A/G-Sepharose beads (GE healthcare, Chicago) were added for precipitating the immune complexes. After 2 hours of rotation, the immune complexes were bathed with a low salt washing buffer followed by washing with high salt washing buffer. Eventually, the immune precipitates were purified and detected by the qRT-PCR assay.
2.10 Fluorescence in situ hybridization analysis
lncRNA FISH assay was applied for observing the presence of OIP5-AS1 in GC cells with the usage of Ribo lncRNA FISH Probe Mix (Red) (C10920; Ribobio, Shanghai, China). First, AGS and SGC7901 cells were immobilized by 4% formaldehyde for 15 minutes followed by PBS washing. Fixed GC cells then underwent protein K digestion and ethanol dehydration. After OIP5-AS1 was captured by Cy3-labeled probes, Cy3-OIP5-AS1 was hybridized in hybridization buffer prepared in advance in air-dried AGS and SGC7901 cells. Subsequently, the slides were washed and dehydrated. At last, the nuclei were stained by 4,6-diamidino-2-phenylindole (DAPI) and immunofluorescence signal was visualized by confocal microscopy.
2.11 Subcellular fractionation assay
The isolation of nuclear fraction from cytosolic fraction was performed with the PARIS Kit (Life Technologies) applied in light of the manufacturer's instructions. First, AGS and SGC7901 cells were gently washed twice with ice-cold PBS and later lysed in ice cold lysis solution. Cell lysate was ice-cultured for 5 minutes. After centrifugation at 4℃, precipitated RNAs isolated from the nucleus or cytoplasm was washed twice with PBS and subjected to qRT-PCR assay. GAPDH and U6 were taken as cytoplasm and nucleus fractionation indicators, respectively. The primer sequence for U6 is listed: forward; 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse; 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
2.12 Statistical analysis
All experiment data processing and analyzing was proceeded using the statistics software SPSS 20.0 (SPSS, Inc, Chicago) and the results of all assays conducted triplicate independently were expressed as the mean value ± standard deviation. Mean values between two groups or among several groups were compared by virtue of Student t-tests or one-way analysis of variance. P < .05 was demarcated as statistically significant.
3 RESULTS
3.1 OIP5-AS1 silencing impairs cell growth and induces promotion of cell apoptosis in gastric cancer
In view of the well-established tumor promoter role of OIP5-AS1 in several malignancies and the fact that the involvement of OIP5-AS1 in GC has not been demonstrated, first, we examined the basal levels of OIP5-AS1 in four GC cell lines (SGC7901, AGS, MKN45, and BGC803) and paired normal gastric epithelial GES-1 cells. The expression of OIP5-AS1 in four GC cell lines clearly surpassed that of GES-1 cells (Figure 1A). AGS and SGC7901 cells, which presented an obviously elevated level of OIP5-AS1 among four GC cell lines, were selected and subjected to the following OIP5-AS1 interference assays. OIP5-AS1 interference efficiency was testified in the qRT-PCR analysis (Figure 1B). sh-OIP5-AS1#1 and sh-OIP5-AS1#2 plasmids were utilized for the subsequent assays to explore the functions of the altered expression of OIP5-AS1 on GC cellular processes. AGS and SGC7901 cells with low OIP5-AS1 expression in the presence of sh-OIP5-AS1#1 or sh-OIP5-AS1#2 grew much slower than those with the transfection of sh-NC as demonstrated by the CCK-8 assay (Figure 1C). Results from flow cytometry apoptosis analysis suggested that OIP5-AS1-silenced GC cells underwent severe apoptosis (Figure 1D). As shown in Figure 1E, the migratory capacity of OIP5-AS1-silenced GC cells was noticeably impeded. Above all, it unmasked OIP5-AS1 as a pro-proliferation and pro-migration factor in GC cells.
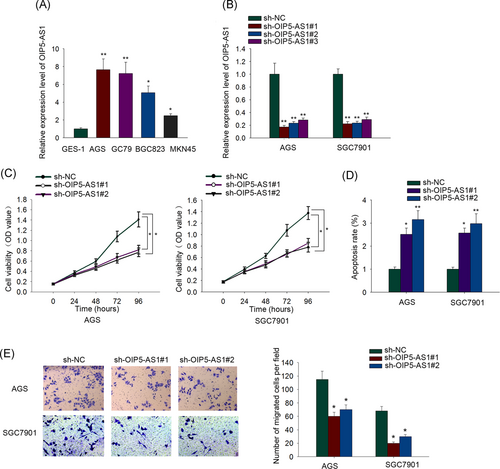
OIP5 antisense RNA 1 (OIP5-AS1) silencing impairs cell growth and induces promotion of cell apoptosis in gastric cancer. A, Real-time quantitative polymerase chain reaction (qRT-PCR) assay revealed a robust upregulation of OIP5-AS1 in gastric cancer (GC) cells relative to normal gastric epithelial GES-1 cells. B, Transfection efficiency of sh-OIP5-AS1#1, sh-OIP5-AS1#2, and sh-OIP5-AS1#3 into AGS and SGC7901 cells was ensured by qRT-PCR. C, AGS and SGC7901 cells after OIP5-AS1 depletion were subjected to cell counting kit-8 (CCK-8) assay. D, Flow cytometry apoptosis analysis evaluated the effect of OIP5-AS1 silencing on AGS and SGC7901 cell apoptosis. E, Transwell assay investigated the impact of OIP5-AS1 knockdown on AGS and SGC7901 cell migratory capacity. *P < .05, **P < .01
3.2 Nuclear OIP5-AS1 interacts with EZH2 and negatively mediates NLRP6 expression in GC cells
To unravel the mechanism that explains how OIP5-AS1 exerted its effect in GC cells, the presence of OIP5-AS1 in GC cells was observed by subcellular fractionation assay and FISH analysis, and higher expression level of OIP5-AS1 was found in the nucleus of AGS and SGC7901 cells vs cytoplasm (Figure 2A,B). Namely, OIP5-AS1 may serve as a critical regulator of the phenotypes of GC cells at a transcriptional level. As presented in previous studies, lncRNAs could recruit polycomb repressive complex 2 (PRC2) which includes EZH2 to downstream target gene promoter, repressing relevant gene expression.24-26 Thus, a reasonable hypothesis was formulated that OIP5-AS1 bound to EZH2, an essential PRC2 subunit, which was validated in the following RIP assay with antibody against EZH2. It uncovered that increased OIP5-AS1 that was successfully immunoprecipitated was detected in anti-EZH2 group not in control immunoglobulin G (IgG) group (Figure 2C). As discussed above, the recruitment of EZH2 mediated by lncRNAs resulted in the inactivation of tumor suppressor genes, which was responsible for tumor progression. That is because the lncRNA-mediated recruitment of EZH2 to promoters of target genes could lead to increased trimethylation of H3K27 on the target gene promoter, and, as a result, inactivate gene transcription. Besides, NLRP6 was previously confirmed to suppress gastric tumorigenesis.27 UCSC Genome Browser (http://genome.ucsc.edu/) and GeneCards webserver (https://www.genecards.org/) were applied and indicated that EZH2 could associate with NLRP6 promoter. Consequently, hypothesis was made that EZH2 recruited by OIP5-AS1 could bind to NLRP6 promoter and suppress NLRP6 expression. As expected, the expression of NLRP6 examined by qRT-PCR analysis in GC cells was remarkably reduced relative to normal GES-1 cells, which was consistent with the predocumented tumor inhibitor role of NLRP6 (Figure 2D). Whether OIP5-AS1 was able to mediate NLRP6 expression was also probed. qRT-PCR analysis and Western blot assay exhibited that silencing OIP5-AS1 notably decreased mRNA and protein levels of NLRP6, confirming the inhibitory impact of OIP5-AS1 on NLRP6 (Figure 2E,F). Altogether, there is an association between OIP5-AS1 and EZH2 and OIP5-AS1 hampered NLRP6 expression.
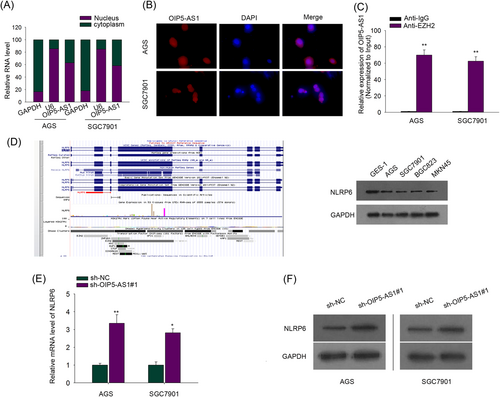
Nuclear OIP5-AS1 interacts with enhancer of zeste homolog 2 (EZH2) and negatively mediates Nod-like receptor pyrin domain-containing protein 6 (NLRP6) expression in GC cells. A, B, Subcellular fractionation assay and FISH analysis demonstrated that OIP5-AS1 was mainly located in the nucleus. C, RIP followed by OIP5-AS1 qPCR analyzed OIP5-AS1 expression endogenously interacted with EZH2. D, Western blot assay assessed the protein content of NLRP6 in GC cells and normal gastric epithelial GES-1 cells. E, qRT-PCR assay examined the NLRP6 messenger RNA (mRNA) level in AGS and SGC7901 cells after transfection with sh-NC or sh-OIP5-AS1#1. F, Western blot assay detected NLRP6 protein level in AGS and SGC7901 cells after transfection with sh-NC or sh-OIP5-AS1#1. *P < .05, **P < .01. GC, gastric cancer; OIP5-AS1, OIP5 antisense RNA 1; qRT-PCR, real-time quantitative polymerase chain reaction
3.3 OIP5-AS1 represses NLRP6 expression by recruiting EZH2, increasing the H3K27me3 enrichment of NLRP6 promoter
To certify whether OIP5-AS1 functioned an inhibitory effect on NLRP6 expression in GC cells by interacting with EZH2, qRT-PCR and Western blot analysis were implemented. NLRP6 mRNA and protein levels drastically declined in the pcDNA3.1/EZH2 group, not in the control group (Figure 3A,B). Besides, ChIP assay was employed to monitor the interaction of EZH2 and NLRP6 promoter region and also verified the binding between them (Figure 3C). Lastly, to probe the effect of altered OIP5-AS1 expression on the binding of EZH2 to NLRP6 promoter region in GC cells, ChIP assay followed by qRT-PCR analysis was implemented and manifested that after OIP5-AS1 blockade, the enrichment of H3K27me3 and occupancy of EZH2 in the NLRP6 promoter region was obviously decreased in GC cells (Figure 3D). These results proved that OIP5-AS1 interfered in the enrichment of H3K27me3 and EZH2 in NLRP6 promoter region, obstructing NLRP6 translation.
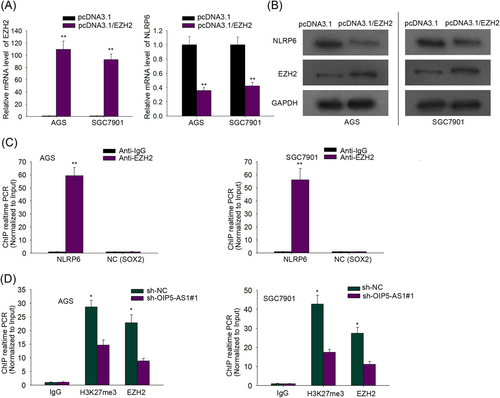
OIP5-AS1 represses NLRP6 expression by recruiting EZH2, increasing the H3K27me3 enrichment of NLRP6 promoter. A, qRT-PCR assay interrogated EZH2 mRNA level after transfection of pcDNA3.1 or pcDNA3.1/EZH2 into AGS and SGC7901 cells. B, NLRP6 protein content was probed by Western blot assay in AGS and SGC7901 cells transfected as indicated. C, Chromatin immunoprecipitation (ChIP) assay suggested that EHZ2 could bind to NLRP6 promoter region in AGS and SGC7901 cells with SOX2 serving as a negative control. *P < .05, **P < .01. D, ChIP assay manifested that OIP5-AS1 inhibition reduced the expression level of EZH2 and H3K27me3 enriched in the NLRP6 promoter region. EZH2, enhancer of zeste homolog 2; mRNA, messenger RNA; NLRP6, Nod-like receptor pyrin domain-containing protein 6; OIP5-AS1, OIP5 antisense RNA 1; qRT-PCR, real-time quantitative polymerase chain reaction
3.4 OIP5-AS1 boosts gastric cell growth and migration through binding EZH2 and epigenetically inhibiting NLRP6 expression
To further validate that OIP5-AS1/EZH2/NLRP6 signaling pathway mediates GC progression in vitro, we silenced NLRP6 in AGS cells and the efficacy was tested by qRT-PCR (Figure 4A). Knockdown of NLRP6 or upregulation of EZH2 restored the inhibited AGS cell proliferation caused by OIP5-AS1 depletion (Figure 4B). EZH2 overexpression or NLRP6 depletion reversed OIP5-AS1 silencing-induced AGS cell apoptosis promotion (Figure 4C). With regarding to cell migration, sh-OIP5-AS1#1-transfected AGS cell demonstrated declined migration capacity, while EZH2 augmentation or NLRP6 inhibition reverted this trend (Figure 4D). Taken together, we attested that the contribution of OIP5-AS1 to GC cell growth and migration was made by its epigenetic regulation of NLRP6 transcription inhibition via interacting with EZH2.

OIP5-AS1 boosts gastric cell growth and migration, through binding EZH2 and epigenetically inhibiting NLRP6 expression. A, The expression of NLRP6 in the presence of sh-NLRP6 was surveyed in AGS cells. B, AGS cell growth was monitored by CCK-8 assay as transfected with indicated plasmids. C, AGS cell apoptosis was evaluated by flow cytometry apoptosis analysis after the transfection of indicated plasmids. D, The migration potential of AGS cells transfected with plasmids as indicated was estimated by transwell assay. *P < .05, **P < .01. CCK-8, cell counting kit-8; EZH2, enhancer of zeste homolog 2; NLRP6, Nod-like receptor pyrin domain-containing protein 6; OIP5-AS1, OIP5 antisense RNA 1
4 DISCUSSION
GC, as one of the most common gastrointestinal tumors, leads to a large amount of cancer-induced mortalities globally and worse still, diagnosis defects and poor prognosis haunt people all the time.12, 28 Although more and more new mechanisms with respect to genetic and epigenetic alterations in GC have been extensively annotated, the risk of poor survival rate of GC patients is frightening. Thus, a drastic improvement in efficiently identifying novel biomarkers might offer a more pragmatic approach for boosting GC prognosis.
lncRNAs, a cluster of actively transcribed ncRNAs, has been broadly proposed to diagnostically or therapeutically work as a newfound biomarker during multiple tumor carcinogenesis and progression. Besides, the determination of their critical roles in cancer development is also dependent on their location in cells, which in other words, access to interaction with different molecules distributed either in the nucleus or cytoplasm provides a flexible mechanism of actions of lncRNAs.29 For instance, the broad tendency of lncRNAs existing in nucleus is to interact with genomic DNA, transcription factors, spliceosomes, chromatin regulators, and so on, implicating in transcriptional and epigenetic regulations containing histone modifications.30 Regulatory ncRNAs such as PCAT6,31 LINC00968,32 TUG1,33 and CASC1534 were reported to interact with EZH2, increasing trimethylation of H3K27 on the target gene promoters. On the one hand, lncRNA OIP5-AS1 involved in this study, a tumor promoter in several cancers,19-23 was not documented to partake in GC oncogenesis. On the other hand, the mechanism of OIP5-AS1-recruited EZH2 regulates tumor proceedings has never been noted either. The cancer-promoting function of OIP5-AS1 was consistently verified in previous studies and in our study. As presented in our work, OIP5-AS1 contributed to GC cell proliferation and migration while impaired apoptosis. Furthermore, OIP5-AS1-regulated epigenetic modification by scaffolding EZH2 was first put forward in the present study.
NLRP6 is a constituent part of the inflammasome, which was initially clarified to protect mice from intestinal injury and colon cancer.35 The vital role played by NLRP6 in innate immunity and host defense was certified.28 Besides, its downregulation and the tumor suppressor role were also uncovered in GC.27 NLRP6 depletion is correlated with a rising incidence of lymph node metastasis and poor prognosis. Anterior research and statistical analysis provide corroborative evidence for the current study. Relative to previous literature, our work first pointed out that the suppressive function of NLRP6 in GC was induced by the enrichment of H3K27me3 on its promoter region through lncRNA recruiting EZH2.
In summary, this study indicated the function of OIP5-AS1 in facilitating GC cell proliferation, migration, and in hindering apoptosis via binding EZH2 and epigenetically repressing NLRP6. The study is expected to shed new insight into GC treatment improvement and more efficient diagnosis.
ACKNOWLEDGMENT
The authors are grateful to all participants in this study.
AUTHOR CONTRIBUTIONS
Yunlei Bai was in charge of article writing and bringing practical experience. Sheng Li was responsible for recording the experience results.