Regulatory effects of lncRNA ATB targeting miR-200c on proliferation and apoptosis of colorectal cancer cells
Abstract
This investigation was intended to elucidate whether long noncoding RNA (lncRNA)-activated by transforming growth factor-β (ATB) interacting with miR-200c could mediate colorectal cancer (CRC) progression, offering potential strategies for diagnosing and treating CRC. Here totally 315 patients with CRC were recruited, and their CRC tissues and adjacent normal tissues were gathered. Concurrently, four colon cancer cell lines (ie, SW620, Lovo, HCT116, and SW480) and the human colon mucosal epithelial cell line (NCM460) were also purchased. Moreover, si-ATB, si-NC, miR-200c mimic, miR-200c inhibitor, and miR-NC were prepared for transfection into the CRC cells, and their effects on CRC cell lines were evaluated based on the conduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, colony formation assay, and flow cytometry assay. Eventually, the Luciferase reporter gene assay was carried out to judge if there existed a targeted relationship between ATB and miR-200c. The results of Cox regression analyses suggested that overexpressed lncRNA ATB, underexpressed miR-200c, poor tumor differentiation, lymph-vascular invasion, and perineural invasion were symbolic of shortened survival of the patients with CRC (all P < .05). Besides, transfection of pcDNA3.1-ATB and miR-200c inhibitor could boost the viability and proliferation of Lovo and SW620 cell lines (all P < .05). Meanwhile, the expressions of p53 and p21 were also reduced under treatments of pcDNA3.1-ATB and miR-200c inhibitor (P < .05). In addition, CDK2 seemed to reverse the contribution of miR-200c to intensifying viability and proliferation of Lovo and SW420 cell lines (P < .05). Furthermore, ATB might downregulate miR-200c expression by targeting it (P < .05), and CDK2 was subjected to dual regulation of both ATB and miR-200c (P < .05). In conclusion, the lncRNA ATB/miR-200c/CDK2 signaling was responsible for intensified proliferation and prohibited apoptosis of CRC cells, which might provide effective approaches for diagnosing and treating CRC.
1 INTRODUCTION
Colorectal cancer (CRC) was one common malignancy arising from the human digestive tract, serving as a major account for cancer-related death.1, 2 Allowing for the indistinct symptoms of early-stage CRC, a majority of the CRC population remained undiagnosed until their advanced stage.3 Moreover, the patients with CRC accompanied by distant metastasis were documented with a 5-year survival of less than 50%, which stated the significance of diagnosing early CRC. Hence, locating novel biomarkers that particularly indicated the pathogenesis of CRC appeared vital for subsequent diagnosis and prognostic estimation among CRC sufferers.4
In accordance with the encyclopedia of DNA elements initiated in 2003, approximately 65% of genes were transcribed into noncoding RNAs (ncRNAs), and the ncRNAs that possessed more than 200 nucleotides were acknowledged as long noncoding RNAs (lncRNAs).5-9 Intriguingly, the lncRNAs were increasingly concerned regarding their modulation of tumorigenesis.10, 11 Taking CRC as the instance, the patients with CRC that carried high expression of lncRNA PVT1 were associated with more aggressive vascular invasion and lymphatic metastasis than those expressed relatively low expression of PVT1.12, 13 Furthermore, overexpressed lncRNA PCAT-1 could symbolize the unfavorable outcome of the patients with CRC,14, 15 and lncRNA MALAT-1 might serve as a facilitator for CRC onset due to its reinforcing proliferation and invasion of CRC cells.16, 17 Apart from the above, lncRNA-activated by transforming growth factor-β (ATB), which was generally activated by TGF-β, was also a reliable candidate for indicating the poor prognosis of the patients with CRC,18 and it served as a potent participant in the pathogenesis of CRC exacerbation.19 Nevertheless, the action mode of lncRNA ATB underlying CRC development was poorly reported.
As formerly documented, the interaction of lncRNA ATB with miR-200 family became pronounced with the progression of certain neoplasms, such as glioma and osteosarcoma.20, 21 Moreover, lncRNA ATB could accelerate the development of esophageal squamous cell carcinoma via dysregulating miR-200b/Kindlin-2 signaling,22 and it was also involved with the onset of keloid by suppression of miR-200c.23 In fact, the miR-200c mentioned here was responsible for disordered actions within various tumors, including gastric cancer, renal clear cell carcinoma, ovarian cancer, and CRC.24-27 Despite the facilitation of miR-200c for CRC progression,28-30 the upstream molecules (eg, lncRNAs) that enabled miR-200c to play such a part remained obscure.
Given the potential linkage of lncRNA ATB with miR-200c and their respective contributions to CRC development, the lncRNA ATB acting on miR-200c could be implicated in the etiology of CRC onset and progression. Thus, the present investigation was intended to elucidate this point, providing evidence for applying lncRNA ATB and miR-200c into diagnosis and treatment of CRC.
2 MATERIALS AND METHODS
2.1 Inclusion of CRC subjects
A total of 315 pairs of CRC tissues and adjacent normal tissues were removed from the patients with CRC who received surgeries in the First Affiliated Hospital of Bengbu Medical College from January 2012 to March 2013. The subjects were pathologically confirmed as CRC, and they hardly underwent any antitumor therapies before their tissues were obtained. Besides, the patients with CRC were followed up from the date when they experienced surgery or pathological biopsy to 1 December 2018. Moreover, this investigation has obtained permission from the First Affiliated Hospital of Bengbu Medical College and the ethics committee of the First Affiliated Hospital of Bengbu Medical College. All the participants have signed informed consents.
2.2 Cell culture
The human colon mucosal epithelial cell line (NCM460) and four colon cancer cell lines, including SW620, Lovo, HCT116, and SW480 were purchased from Shanghai Institute of Biochemistry and Cell Biology that was affiliated to Chinese Academy of Sciences (Shanghai, China). The cells were cultured within Dulbecco's modified Eagle's medium that contained 10% fetal bovine serum (FBS; Life Technologies) and 100 U/mL penicillin/streptomycin. The cultivation atmosphere was maintained as 5% CO2, saturated humidity and 37°C. When cell confluence reached about 80%, the cells were trypsinized into single-cell suspension and were passaged at the ratio of 1:3.
2.3 Cell transfection
Around 5 × 105 CRC cells were seeded into six-well culture plates, and they were prepared for transfection after 24-hour culture. Subsequently, si-ATB (si-ATB-1: sense: 5′-TATGGCCTAGATTACCTTTCCATT-3′, antisense: 5′-TGGAAAGGTAATCTAGGCCATATT-3′, GenePharma, China; si-ATB-2: 5′-GTCTGTATTTGCGAATACCTTT-3′, antisense: 5′-AAAGGTATTCGCAAATACAGAC-3′, GenePharma), si-NC (sense: 5′-GATCCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTG-3′, antisense: 5′-AATTCAAAAAAGTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAACG-3′, GenePharma), miR-200c mimic (5′-UAAUACUGCCGGGUAAUGAUGGA-3′, Invitrogen), miR-200c inhibitor (5′-UCCAUCAUUACCCGGCAGUAUUA-3′, Invitrogen) and miR-NC (5′-GUUCAUCAUGGCGAUGAGAGUAA-3′, Invitrogen) were transfected into CRC cells according to the instructions of Lipofectamine 2000 kit (Invitrogen). The culture solution was replaced with the medium that contained 10% FBS, and cells were harvested 48 hours later.
2.4 Implementation of reverse transcription-polymerase chain reaction
After the 48-hour culture, the total RNA was extracted based on the phenol-trichloromethane method. The integrity of the extracted RNAs was judged through performing agarose gel electrophoresis and UV spectrophotometry, and it was deemed as qualified when the absorbance ratio of 260 and 280 nm lied within the range of 1.8 to 2.1. Subsequently, the total RNA was reversely transcribed into complementary DNAs (cDNAs) in light of the guidance of PrimeScript RT Reagent Kit (TaKaRa, Japan), under conditions of 37°C for 30 minutes and 98°C for 5 minutes. With primers shown in Table 1 (Sangon, China), the cDNAs were then amplified via the implementation of polymerase chain reaction (TaKaRa). Besides, the reaction conditions for ATB and GAPDH were summarized as (a) predegeneration at 95°C for 30 seconds and (b) 40 cycles of degeneration at 95°C for 5 seconds and annealing at 60°C for 30 seconds, and the reaction procedures of miR-200c and U6 were particularized as (a) predegeneration at 95°C for 20 seconds and (b) 40 cycles of degeneration at 94°C for 5 seconds, annealing at 55°C for 20 seconds and extension at 72°C for 20 seconds. The expressions of lncRNA ATB and miR-200c were quantified by consulting method,31 with GAPDH and U6, respectively, as the internal reference.
| Genes | Sequences (5′-3′) |
|---|---|
| lncRNA ATB | ACAAGCTGTGCAGTCTCAGG (forward) |
| CTAGGCCCAAAGACAATGGA (reverse) | |
| miR-200c | TACATCATAATACTGCCGGGTAA (forward) |
| GGATTGGATGTTCTCCACAGTCTC (reverse) | |
| GAPDH | AGCAAGAGCACAAGAGGAAG (forward) |
| GGTTGAGCACAGGGTACTTT (reverse) | |
| U6 | ATTGGAACGATACAGAGAAGATT (forward) |
| GGAACGCTTCACGAATTTG (reverse) |
- Abbreviation: lncRNA ATB, long noncoding RNA-activated by transforming growth factor-β.
2.5 Western blotting
The total protein was extracted after addition of radioimmune-precipitation assay lysate, which was composed of 50 mmol/L Tris (pH8.0), 150 mmol/L NaCl, 1 mmol/L ethylenediaminetetraacetic acid, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS) and proteinase inhibitor. The concentration of the extracted proteins was determined by means of bisquinoline carboxylic acid (BCA) method. Next, 40 μg protein for each well was managed for conduction of 10% SDS-polyacrylamide gel electrophoresis, which was then transferred onto the polyvinylidene fluoride membrane on the basis of the wet transfer. After 2-hour blockage with 5% bovine serum albumin at room temperature, we added the diluted rabbit-anti-human primary antibodies (Abcam) against p53 (1:1000, catalog no.: ab32389), p21 (1:2000, catalog no.: ab109520), CDK2 (1:1000, catalog no.: ab32147), and GAPDH (1:10000, catalog no.: ab181602) for overnight incubation at 4°C. The samples were then washed three times with polysorbate and triethanolamine-buffered saline solution (TBST), and each time lasted for 15 minutes. Subsequently, the goat anti-rabbit secondary antibodies marked by horseradish peroxidase (1:3000, catalog no.: ab6721, Abcam) were supplemented for another 1-hour incubation at room temperature. After rinsing cells with TBST three times, the enhanced chemiluminescence (plusECL) luminescence reagent (Amersham Biosciences) was added for the development. With β-actin as the reference, the Chemi Imager 5500 V2.03 software (Kadak) was applied to scan the test results, and the image analysis system (Fluor Chen 2.0; Kadak) was used to quantitate the proteins.
2.6 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
After 48-hour transfection, the cells in the logarithmic phase were digested through the usage of trypsin, and 200 μL cell suspension (3 × 104 cells/mL) was then added into each well. After cultivation of the cells for 1 to 4 days, 20 μL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (5 mg/mL; Promega) was supplemented into each well. The culture medium was discarded, and each well was added with 150 μL dimethyl sulfoxide to terminate the reaction. At last, the optical density values of each well were determined at the wavelength of 490 nm on the microplate reader (Thermo Fisher Scientific).
2.7 Colony formation assay
The cells were seeded into six-well plates at the density of 200 per well, and were then cultivated in 5% CO2 at 37°C for 2 weeks. At the time that macroscopic colony was observed, the culturing process was finalized. Subsequently, the cells were fixed with 4% paraformaldehyde for 10 minutes, followed by being stained with Giemsa for 20 minutes. Finally, cells after air-drying were counted.
2.8 Flow cytometry
The cells transfected for 48 hours were prepared into single-cell suspensions, and then they were centrifuged at 1200 rpm for 5 minutes. The acquired supernatants were discarded, and the cells were rinsed with precooled phosphate-buffered saline for twice. With 1X binding buffer (500 μL) added to resuspend the cells, 5 μL fluorescein isothiocyanate-labeled Annexin V (Beckton Dickinson) was afterward supplemented to mark the cells. After supplementation of 10 μL propidium iodide, the cells were incubated at room temperature for 5 minutes, immediately after which the cells were detected utilizing flow cytometry (model: FACScan; Beckton Dickinson).
2.9 Luciferase reporter gene assay
The Genepharma company was entrusted to construct fluorescent reporter vectors of pmirGLO (Promega)-ATB-wild-type (wt), pmirGLO-CDK2-wt, pmirGLO-ATB-mutant (mut), and pmirGLO-CDK2-mut. Then the obtained pmirGLO-ATB-wt, pmirGLO-CDK2-wt, pmirGLO-ATB-mut, and pmirGLO-CDK2-mut were, respectively, cotransfected with miR-200c mimic and miR-NC into the CRC cell lines. Forty-eight hours later, the firefly luciferase and Renilla luciferase activities of the cells were examined, according to the specifications of the Dual-Luciferase Activity Assay Kit (Promega).
2.10 Statistical analyses
All the statistical analyses were carried out through the usage of SPSS v. 19.0 software. The measurement data expressed as mean ± standard deviation were compared based on the student t test and one-way analysis of variance (ANOVA), when they conformed to normal distribution and homoscedasticity. Otherwise, the Kruska-Wallis H test was adopted for the comparisons. Moreover, the associations of genetic expressions with clinical-pathological parameters of the patients with CRC were evaluated by virtue of the χ2 test. Furthermore, whether genetic expressions were associated with the prognosis of the patients with CRC was analyzed through devising Kaplan-Meier curves, and the Cox proportional hazard model was established to figure out factors that could significantly affect the prognosis of patients with CRC. The differences would be deemed as statistically significant in case of P < .05.
3 RESULTS
3.1 lncRNA ATB and miR-200c were associated with clinical features of the patients with CRC
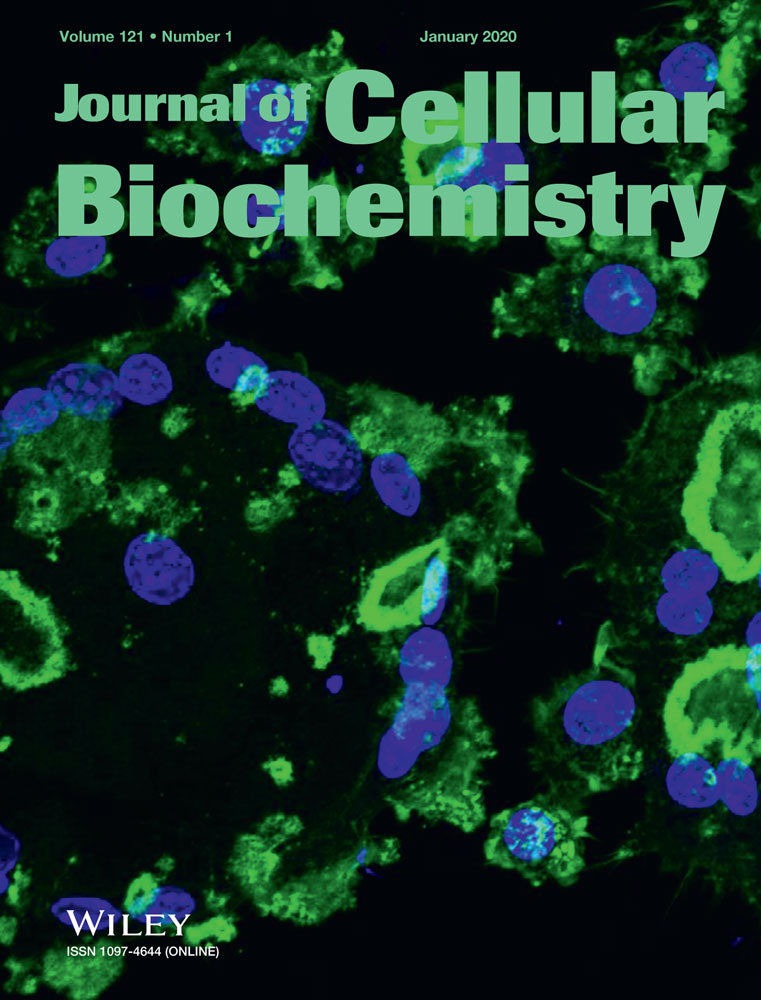
Among the recruited patients with CRC, their CRC tissues exhibited higher ATB expression and lower miR-200c expression than adjacent nontumor tissues (P < .05) (Figure 1A), and an inverse correlation was discovered between the ATB expression and the miR-200c expressions (rs = −0.414, P < .001; Figure 1B). In addition, with the median expression of lncRNA ATB and miR-200c as the dividing point, the 316 patients with CRC were, respectively, split into highly expressed lncRNA ATB group (n = 200) and lowly expressed lncRNA ATB group (n = 116), as well as highly expressed miR-200c group (n = 124) and lowly expressed miR-200c group (n = 192; Table 2). It was manifested that highly expressed lncRNA ATB and lowly expressed miR-200c were more intensively determined within CRC population whose traits consisted of poor tumor differentiation, lymph-vascular invasion, perineural invasion, and advanced stage of the tumor, node, metastasis (TNM) staging system (all P < .05). The multivariate analyses further supported that overexpressed lncRNA ATB (hazard ratio [HR] = 1.89, 95% confidence interval [95% CI] = 1.12-3.13), underexpressed miR-200c (HR = 2.04, 95% CI = 1.23-3.40), poor tumor differentiation (HR = 1.69, 95% CI = 1.02-2.86), invasion of lymph-vascular (HR = 2.50, 95% CI = 1.45-4.35) and perineural invasion (HR = 1.92, 95% CI = 1.11-3.23) could act as independent predictors for the adverse outcome of the patients with CRC (Table 3). The survival curve also vividly demonstrated a significantly shortened survival of patients with CRC who carried overexpressed lncRNA ATB or underexpressed miR-200c, when compared with ones carrying underexpressed lncRNA ATB and overexpressed miR-200c (P < .001; Figure 1C).
Association of lncRNA ATB and miR-200c expressions with the prognosis of patients with colorectal cancer. A, The expressions of lncRNA ATB and miR-200c were compared between colorectal cancer tissues and adjacent nontumor tissues. *P < .05 when compared with adjacent nontumor tissues. B, The lncRNA ATB was inversely correlated with miR-200c expressions among 316 patients recruited with colorectal cancer. C, The lowly expressed lncRNA ATB and the highly expressed miR-200c could serve as the protective parameters for the favorable prognosis of patients with colorectal cancer. lncRNA ATB, long noncoding RNA-activated by transforming growth factor-β
| lncRNA ATB expression | miR-200c expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinical features N = 316 | Low | High | χ2 | P value | Low | High | χ2 | P value |
| Age, y | ||||||||
| <65 | 84 | 128 | 134 | 78 | ||||
| ≥65 | 32 | 72 | 2.35 | .125 | 58 | 46 | 1.62 | .203 |
| Sex | ||||||||
| Male | 76 | 116 | 121 | 71 | ||||
| Female | 40 | 84 | 1.74 | .187 | 71 | 53 | 1.05 | .306 |
| Tumor site | ||||||||
| Rectum | 59 | 115 | 100 | 74 | ||||
| Colon | 57 | 85 | 1.31 | .253 | 92 | 50 | 1.76 | .185 |
| Tumor differentiation | ||||||||
| Well/moderate | 84 | 119 | 113 | 90 | ||||
| Poor | 32 | 81 | 5.33 | .021 | 79 | 34 | 6.18 | .013 |
| Tumor size, mm | ||||||||
| <30 | 83 | 132 | 123 | 92 | ||||
| ≥30 | 33 | 68 | 1.04 | .308 | 69 | 32 | 3.56 | .059 |
| Lymph vascular invasion | ||||||||
| Absence | 94 | 130 | 127 | 97 | ||||
| Presence | 22 | 70 | 9.15 | .003 | 65 | 27 | 5.33 | .021 |
| Perineural invasion | ||||||||
| Absence | 89 | 131 | 122 | 98 | ||||
| Presence | 27 | 69 | 4.37 | .037 | 70 | 26 | 8.55 | .004 |
| TNM stage | ||||||||
| Stage II | 57 | 70 | 68 | 59 | ||||
| Stage III | 59 | 130 | 6.11 | .014 | 124 | 65 | 4.64 | .031 |
- Abbreviations: lncRNA ATB, long noncoding RNA-activated by transforming growth factor-β; TNM, tumor, node, metastasis.
| Clinical features | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| ATB expression | ||||||
| High vs low | 2.50 | 1.56-4.00 | <.001 | 1.89 | 1.12-3.13 | .016 |
| miR-200c expression | ||||||
| Low vs high | 2.61 | 1.64-4.16 | <.001 | 2.04 | 1.23-3.40 | .006 |
| Age, y | ||||||
| <65 vs ≥65 | 1.08 | 0.67-1.72 | .755 | 1.24 | 0.74-2.09 | .419 |
| Sex | ||||||
| Male vs female | 0.94 | 0.60-1.47 | .773 | 0.88 | 0.53-1.44 | .599 |
| Tumor site | ||||||
| Rectum vs colon | 1.06 | 0.68-1.65 | .795 | 1.18 | 0.72-1.93 | .509 |
| Tumor differentiation | ||||||
| Poor vs well/moderate | 1.85 | 1.16-2.94 | .01 | 1.69 | 1.02-2.86 | .042 |
| Tumor size, mm | ||||||
| ≥5 vs <5 | 1.08 | 0.67-1.72 | .76 | 0.81 | 0.48-1.37 | .434 |
| Lymph vascular invasion | ||||||
| Presence vs absence | 2.78 | 1.67-4.76 | <.001 | 2.50 | 1.45-4.35 | .001 |
| Perineural invasion | ||||||
| Presence vs absence | 2.08 | 1.27-3.45 | .004 | 1.92 | 1.11-3.23 | .018 |
| TNM stage | ||||||
| III vs II | 0.96 | 0.61-1.52 | .875 | 0.81 | 0.49-1.35 | .422 |
- Abbreviations: 95% CI, 95% confidence interval; ATB, activated by transforming growth factor-β; TNM, tumor, node, metastasis.
3.2 Contribution of lncRNA ATB and miR-200c to proliferation and apoptosis of CRC cells
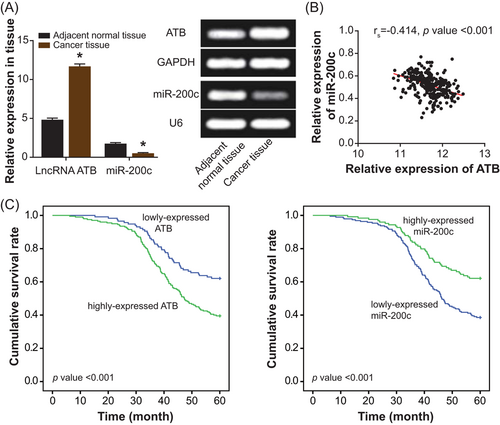
The SW620, Lovo, HCT116, and SW480 cell lines were detected with incremental ATB expression and lessened miR-200c expression in comparison with NCM460 cell line (P < .05; Figure 2A). Due to that Lovo and SW620 cell lines displayed more pronounced expressional change of ATB and miR-200c than NCM460 cell line (P < .05; Figure 2A), they were chosen to conduct the following cellular experiments. After being transfected with si-ATB-1 and si-ATB-2, the Lovo and SW620 cell lines were measured with an observably depressed expression of ATB (P < .05; Figure 2B). Conversely, the ATB expression was enhanced within the CRC cell lines under the transfection of pcDNA3.1-ATB (P < .05; Figure 2B). With regard to miR-200c, its expression was increased and decreased after respective treatments of miR-200c mimic and miR-200c inhibitor (P < .05; Figure 2C).
Contribution of lncRNA ATB and miR-200c to proliferation and apoptosis of colorectal cancer cell lines. A, The expression of lncRNA ATB was compared among SW620, Lovo, HCT116, SW480, and NCM460 cell lines. *P < .05 when compared with NCM460 cell line. B, The expression of lncRNA ATB within Lovo and SW620 cell lines was detected after treatments of NC, pcDNA3.1-ATB, si-ATB-1, and si-ATB-2. *P < .05 when compared with NC. C, The expression of miR-200c within Lovo and SW620 cell lines was determined among miR-NC, miR-200c mimic, and miR-200c inhibitor groups. D, The viability of Lovo and SW620 cell lines was compared among NC, pcDNA3.1-ATB, si-ATB-1, miR-200c mimic, and miR-200c inhibitor groups. *P < .05 when compared with NC. E, The proliferative intensity of Lovo and SW620 cell lines was evaluated under treatments of NC, pcDNA3.1-ATB, si-ATB-1, miR-200c mimic, and miR-200c inhibitor. *P < .05 when compared with NC. F, The apoptotic percentage of Lovo and SW620 cell lines was appraised among NC, pcDNA3.1-ATB, si-ATB-1, miR-200c mimic, and miR-200c inhibitor groups. *P < .05 when compared with NC. G, Expressions of p21 and p53 were compared within Lovo and SW620 cell lines treated by NC, pcDNA3.1-ATB, si-ATB-1, miR-200c mimic, and miR-200c inhibitor. *P < .05 when compared with NC. FITC, fluorescein isothiocyanate; lncRNA ATB, long noncoding RNA-activated by transforming growth factor-β; NC, negative control; PI, propidium iodide
Furthermore, the viability of Lovo and SW620 cell lines in the pcDNA3.1-ATB group and miR-200c inhibitor group was boosted when compared with NC group (P < .05), while inversely the cell lines in the si-ATB-1 group and miR-200c mimic group were determined with weakened viability (P < .05) (Figure 2D). Similarly, the proliferative strength of Lovo and SW620 cell lines was enhanced under treatments of pcDNA3.1-ATB and miR-200c inhibitor (P < .05), yet it was impaired after transfection of si-ATB-1 and miR-200c mimic (P < .05) (Figure 2E). Correspondingly, the apoptotic percentage of Lovo and SW620 cell lines markedly went up in the si-ATB-1 and miR-200c mimic groups and strikingly declined in the pcDNA3.1-ATB and miR-200c inhibitor groups, with NC group as the control (P < .05) (Figure 2F). To further explain the variation of cell proliferation, we also detected the expressions of p53 and p21, two biomarkers for apoptosis of CRC cells.32 It was derived that the expressions of p53 and p21 were reduced in the pcDNA3.1-ATB and miR-200c inhibitor groups (P < .05), yet they were oppositely heightened within cells of the si-ATB-1 group and miR-200c mimic group (P < .05) (Figure 2G).
3.3 The potential correlation between lncRNA ATB with miR-200c
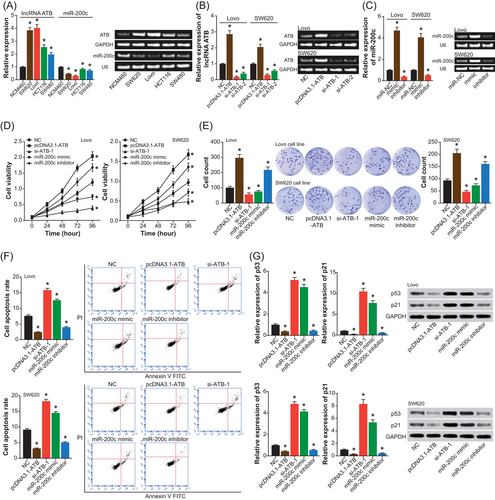
As shown in Figure 3A, hardly any changes in the luciferase activity were observed within Lovo and SW620 cell lines, when mut-type ATB and miR-200c mimic were cotransfected (P > .05). Nonetheless, cotransfection of miR-200c mimic and wt-type ATB led to a significant decrease in the luciferase activity of cell lines (P < .05) (Figure 3A). In addition, the expression of ATB was nearly unaltered, despite the transfection of miR-200c mimic and miR-200c inhibitor (P > .05). On the contrary, transfection of pcDNA3.1-ATB markedly downregulated the expression of miR-200c (P < .05), and silencing of ATB resulted in an upregulation of miR-200c expression (P < .05) (Figure 3B,C).
The targeted regulation of lncRNA ATB on miR-200c within Lovo and SW620 cell lines. A, lncRNA ATB sponged miR-200c in certain sites, and the luciferase activity of Lovo and SW620 cell lines was compared among pmiRGLO-ATB0-wt+miR-200c mimic, pmiRGLO-ATB-mut+miR-200c mimic, and pmiRGLO-ATB-wt+miR-NC groups. *P < .05 when compared with pmiRGLO-ATB-wt+miR-NC. B, The expression of lncRNA ATB was estimated after transfections of miR-NC, miR-200c mimic, and miR-200c inhibitor. *P < .05 when compared with miR-NC. C, The expression of miR-200c was detected within cells under treatments of NC, pcDNA3.1-ATB, and si-ATB-1. *P < .05 when compared with NC. lncRNA ATB, long noncoding RNA-activated by transforming growth factor-β; mut, mutant; NC, negative control; wt, wild-type
3.4 CDK2 mediated the effects of ATB and miR-200c on the proliferation and apoptosis of CRC cells
The CDK2 was expressed distinctly between CRC tissues and adjacent nontumor tissues (P < .05) (Figure 4A), and its expression within CRC cell lines (ie, SW620, Lovo, HCT116, and SW480) was also beyond that within NCM460 cell line (P < .05) (Figure 4B). As was described by Figure 4C, pcDNA3.1-ATB and miR-200c inhibitor produced an increase in the expression of CDK2, yet the expressional intensity of CDK2 was abated within cells of si-ATB-1 and miR-200c mimic groups (P < .05). Besides, CDK2 expression was prominently downregulated and upregulated after respective transfections of si-CDK2 and pcDNA3.1-CDK2 (P < .05) (Figure 4D). And the luciferase activity of pmiRGLO-CDK2-wt+miR-200c mimic group was remarkably below that of pmiRGLO-CDK2-mut+miR-200c mimic group and pmiRGLO-CDK2-wt+miR-NC group (both P < .05), implying a targeted relationship between miR-200c and CDK2 within Lovo and SW420 cell lines (Figure 4E)
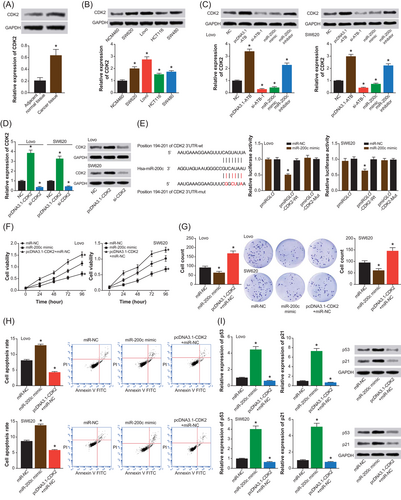
The mediation of CDK2 to modify effects of lncRNA ATB and miR-200c on proliferation and apoptosis of Lovo and SW620 cell lines. A, The CDK2 expression was assessed between colorectal cancer tissues and adjacent nontumor tissues. *P < .05 when compared with adjacent nontumor tissues. B, The expression of CDK2 was compared among SW620, Lovo, HCT116, SW480, and NCM460 cell lines. *P < .05 when compared with NCM460 cell line. C, The CDK2 expression within Lovo and SW620 cell lines was determined among NC, pcDNA3.1-ATB, si-ATB-1, miR-200c mimic, and miR-200c inhibitor groups. *P < .05 when compared with NC. D, The CDK2 expression was obtained from Lovo and SW620 cell lines transfected with pcDNA3.1-CDK2 and si-CDK2. *P < .05 when compared with NC. E, The CDK2 was targeted by miR-200c in certain sites, and the luciferase activity of Lovo and SW620 cell lines was compared among pmiRGLO-CDK2-wt+miR-200c mimic, pmiRGLO-CDK2-mut+miR-200c mimic and pmiRGLO-CDK2-wt+miR-NC groups. *P < .05 when compared with pmiRGLO-CDK2-wt+miR-NC. F-H, The viability, proliferation, and apoptosis of Lovo and SW620 cell lines were measured among groups of miR-NC, miR-200c mimic, and pcDNA3.1-CDK2+miR-NC. *P < .05 when compared with miR-NC. I, The protein levels of p21 and p53 were detected within Lovo and SW620 cell lines among miR-NC, miR-200c mimic, and pcDNA3.1-CDK2+miR-NC groups. *P < .05 when compared with miR-NC. FITC, fluorescein isothiocyanate; lncRNA ATB, long noncoding RNA-activated by transforming growth factor-β; mut, mutant; NC, negative control; PI, propidium iodide; wt, wild-type
In addition, overexpression of CDK2 (ie, pcDNA3.1-CDK2+miR-NC group) seemed to reverse the original inhibitory effects of miR-200c (ie, miR-NC group) on the viability of Lovo and SW420 cell lines (P < .05) (Figure 4F). Furthermore, the proliferation of Lovo and SW420 cell lines was also facilitated in the pcDNA3.1-CDK2+miR-NC group, when compared with miR-NC group and miR-200c mimic group (P < .05) (Figure 4G). On the contrary, the apoptosis of CRC cells was suppressed in the pcDNA3.1-CDK2+miR-NC group, as relative to miR-NC group and miR-200c mimic group (P < .05) (Figure 4H). The p21 and p53 expressions also assumed lower expressions within cells in pcDNA3.1-CDK2+miR-NC group than within cells in either miR-NC group or miR-200c mimic group (P < .05) (Figure 4I).
4 DISCUSSION
The occurrence of CRC, which occurred within the colonic epithelium, could be ascribed to the joint effects of environment and heredity,33 and its early-stage diagnosis and treatment has turned as a challenge that demanded prompt settlement. In response, timely detection of ideal biomarkers for early-stage CRC assumed rising significances, and in-depth investigation of genes underlying CRC pathogenesis could be of help.34-36
Mounting evidence have been indicating that the genesis of CRC was involved with epigenetic modification (eg, methylation),37 modulation of tumor-susceptible genes (eg, p53 and APC),38 and regulation of ncRNAs,39 and CRC was readily induced when the balance of oncogenes and anti-oncogenes was struck.40 For instance, lncRNAs XLOC_006844, LOC152578, and XLOC_000303 were abnormally overexpressed within patients with CRC in comparison with healthy controls, suggesting their potential to facilitate the deterioration of CRC.41 As for lncRNA ATB investigated here, besides CRC, its aberrant overexpression was also visible within pancreatic cancer, CRC, and hepatocellular carcinoma.18, 42, 43 Within this study, lncRNA ATB revealed a conspicuous relevance to the unfavorable outcome of patients with CRC (Figure 1C), and this clinical linkage could be attributed to the role of lncRNA ATB in altering proliferation and apoptosis of CRC cells (Figures 2D and 2F). In addition, we also observed that overexpression of lncRNA ATB could efficiently downregulate expressions of p21 and p53 (Figure 2G). The p21, which belonged to the cyclin-dependent kinase inhibitor family,44 was able to negatively influence proliferation and differentiation of tumor cells,45 and the p53 was also capable of hindering proliferation and invasion of tumor cells.46 All these evidence could deepen the cognition that lncRNA ATB was responsible for CRC development via increasing proliferation and decreasing apoptosis of CRC cells.
Besides, lncRNA ATB sponging microRNAs (miRNAs) has been estimated as a pivotal approach to drive neoplastic progression. For instance, lncRNA ATB might promote tumor invasion and migration by acting on miR-200 and thereby elevating the expression of ZEB1, which was an epithelial-mesenchymal transition (EMT)–specific transcription factor.21 Here we implied miR-200c as the anti-oncogene for CRC, which was subjected to the negative modulation of lncRNA ATB (Figure 3C). Virtually, the expression of miR-200c was obviously upregulated within neoplastic cells of epithelial origin, in comparison with stromal tumor cells, implying the availability of miR-200c in differentiating epithelial tumors from stromal tumors.47 More than that, the unusual underexpression of miR-200c within neoplasms, including gastric cancer, renal clear cell tumor, meningiomas, pituitary tumor, and breast cancer48 also emphasized the protective impacts of miR-200c against neoplasms. Thus, agreements were reached about the antitumor effect of miR-200c, which could be achieved by its disturbing proliferation and accelerating apoptosis of tumor cells (Figures 2D and 2F).
In addition, so far the functional mechanisms of miRNAs were most relevant to their identifying 3′-untranslated region (3′-UTR) of messenger RNAs (mRNAs) based on the principle of complementary base pairing, which could ultimately degrade mRNAs and/or restrain translation of the mRNAs.49 The regulatory effects of miRNAs on specific genes have been supposed as a pivotal contributor to neoplastic aggravation. For example, miR-650 was found to block ING4 expression by acting on the 3′-UTR of ING4 mRNA, which finally promoted tumorigenesis.27 Moreover, miR-195 could directly target E2F3, CDK6, and cyclin D1 so as to procrastinate the proliferative rate of hepatoma carcinoma cells.50 Here we carried out the Luciferase reporter gene assay and confirmed that there existed a targeted relationship between miR-200c and CDK2 (Figure 4E). In addition, the protein level of CDK2 was significantly reduced when miR-200c was overexpressed (Figure 4C), insinuating that miR-200c was capable of modifying the posttranscriptional level of CDK2 by directly targeting it. Actually, the CDK2 was a critical biomarker for improved cell proliferation, considering its synthesizing with cyclin A and cyclin E to control cell cycle51 and its activating c-myc to slow down cell apoptosis.52, 53 Consistently, the oncogenic action of CDK2, whose expression was modified by lncRNA ATB and miR-200c (Figure S1), was also verified within CRC cells (Figure 4C).
Summing up the above, this investigation was advantageous in constructing a molecular axis, namely, lncRNA ATB/miR-200c/CDK2 signaling, that could partly explain the intensified proliferation and prohibited apoptosis of CRC cells, which potentially contributed to poor prognosis of patients with CRC. However, a couple of shortcomings should be remedied in the future. For one thing, lncRNA ATB and miR-200c could, respectively, possess targets of diverse miRNAs and mRNAs, and discovery of multiple miRNAs/genes regulated by a specific lncRNA/miRNA might be conducive to thorough suppression of neoplastic growth. For another, other tumor-related pathogenesis (eg, EMT) should also be explored,54-59 which could assist in more effective control of tumor development. All in all, the above-mentioned disadvantages should be improved in the future.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.