lncRNA ZFPM2-AS1 promotes proliferation via miR-18b-5p/VMA21 axis in lung adenocarcinoma
Min Xue and Weimin Tao are co-first authors.
Abstract
Lung cancer has been proved to be one of the most common kinds of cancers around the globe. Meanwhile, as the predominant type of lung cancer, lung adenocarcinoma (LUAD) has received increasing attention in cancer research. Long noncoding RNAs (lncRNAs) are known to be associated with oncogenesis and progression of various cancers. However, many lncRNAs have not been thoroughly detected in LUAD. In this study, through bioinformatics analysis we found that zinc finger protein multitype 2 antisense RNA 1 (ZFPM2-AS1) was associated with poor prognosis of LUAD patients. Also, ZFPM2-AS1 was detected to be overexpressed in LUAD tissues and cells. Furthermore, ZFPM2-AS1 could promote the proliferation of LUAD cells. Next, miR-18b-5p was found to bind with and negatively regulated by ZFPM2-AS1. VMA21, target gene of miR-18b-5p, could bind with and be negatively regulated by miR-18b-5p. More importantly, both ZFPM2-AS1 and VMA21 were found to be attached to the RNA-induced silencing complex constructed from miR-18b-5p and Ago2. Also, ZFPM2-AS1 could regulate the expression of VMA21. Therefore, ZFPM2-AS1 were confirmed to regulate VMA21 by competitively binding with miR-18b-5p. Finally, rescue assays confirmed that ZFPM2-AS1 could regulate LUAD cell proliferation via miR-18b-5p/VMA21 axis.
1 INTRODUCTION
As one of the most frequently diagnosed cancers, lung cancer has received more and more attention.1 Due to the coaction of atmosphere pollution, tobacco and aging, lung cancer patients are increasing rapidly, causing more than 6000 deaths in China alone each year.2, 3 Lung adenocarcinoma (LUAD) is one subtype of lung cancer, with a 5-year survival rate of merely 15%.4 Despite the progress in medical condition and technology, the prognosis of LUAD patients is far from satisfactory. Therefore, the research on the underlying mechanism of LUAD appears crucial and indispensable.
The progress in long noncoding RNA (lncRNA) might shed a light for cancer researchers. lncRNAs are a cohort of RNAs which cannot be translated into proteins.5 lncRNAs take up more than 98 percent of transcriptomes and are closely related to a series of biological functions of cells.5, 6 Studies have found the dysregulation of lncRNA is closely associated with oncogenesis and progression of various cancers. For example, TINCR were found to suppress proliferation and invasion in lung cancer.7 Or, MCM3AP-AS1 promotes hepatocellular carcinoma progression.8 Also, ZFAS1 could promote bladder cancer tumorigenesis.9
Zinc finger protein multitype 2 antisense RNA 1 (ZFPM2-AS1) is a novel lncRNA which has not received much attention. Before this study, ZFPM2-AS1 were only found to attenuate the p53 pathway and promote gastric carcinogenesis.10 In this study, through bioinformatics analysis we found that ZFPM2-AS1 was associated with poor prognosis of LUAD patients. Also, ZFPM2-AS1 were found to be overexpressed in LUAD tissues and cells. Furthermore, ZFPM2-AS1 could promote the proliferation of LUAD cells. Next, miR-18b-5p was found to bind with and negatively regulated by ZFPM2-AS1. VMA21, target gene of miR-18b-5p, could bind with and was negatively regulated by miR-18b-5p. More importantly, both ZFPM2-AS1 and VMA21 were found to be attached to the RNA-induced silencing complexes (RISCs) constructed by miR-18b-5p and Ago2. Also, ZFPM2-AS1 could regulate the expression of VMA21. Therefore, ZFPM2-AS1 was confirmed to regulate VMA21 by competitively binding with miR-18b-5p. Finally, rescue assays confirmed that ZFPM2-AS1 could regulate LUAD cell proliferation via miR-18b-5p/VMA21 axis.
The ZFPM2-AS1/miR-18b-5p/VMA21 axis is brand new. lncRNA ZFPM2-AS1 might serve as a cutting point in LUAD research, or even a potential target in LUAD treatment in the future.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
LUAD cell lines (A549, H1299, H1260, and H520) and human bronchus epithelial cell (16HBE) were all obtained from the American Type Culture Collection (ATCC; Manassas, VA). All cells were cultured in Roswell Park Memorial Institute 1640 medium (KeyGEN; Nanjing, Jiangsu, China), supplemented with 10% fetal bovine serum, 100 mg/mL streptomycin and 100 U/mL penicillin. All cells were cultured in a humid atmosphere containing 5% CO2 at 37°C.
2.2 Cell transfection
For overexpression, ZFPM2-AS1 and VMA21 were cloned into pcDNA3.1 vectors (Addgene, Cambridge, MA). ShRNAs against ZFPM2-AS1(sh-ZFPM2-AS1#1/2/3) were purchased from Realgene (Nanjing, Jiangsu, China). MiR-18b-5p mimics and NC mimics were obtained from Ambion (GeneCopoeia, Guangzhou, Guangdong, China). All plasmids were transfected into A549 and H1299 cells with Lipofectamine 2000 reagent (Invitrogen, Waltham, MA).
2.3 Quantitative reverse transcription polymerase chain reaction
Total RNA was extracted from cells by TRIzol reagent (Invitrogen). RNAs were reversely transcribed into cDNAs with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster, CA) or TaqMan miRNA Reverse Transcription Kit (Applied Biosystems). The expression levels of ZFPM2-AS1, miR-18b-5p, and VMA21 mRNA were quantified with SYBR Premix Ex Taq (Takara, Dalian, Liaoning, China) on a Roche LightCycler 480 Real-Time PCR System (Applied Biosystems). U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were applied as internal references. The relative expressions were calculated through method. The primer sequences were listed below: ZFPM2-AS1, 5′-TTTCCTACAATGAATCCACCAG-3′ (forward) and 5′-TTTGAGCCACTCTTTGAGG-3′ (reverse); miR-19b-5p, 5′-TGTGCAAATCCATGCAAAACTGA-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse); VMA21, 5′-GCATCTACACTGAAGACGCTCC-3′ (forward) and 5′-GACCACTGCAACAATAGCAGCG-3′ (reverse); GAPDH, 5′-TATGATGATATCAAGAGGGTAGT-3′ (forward) and 5′-TGTATCCAAACTCATTGTCATAC-3′ (reverse); U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse).
2.4 Western blot analysis
Total protein was extracted with radioimmunoprecipitation assay lysis buffer (Beyotime, China). Extracted proteins were first mixed with loading buffer, separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride (PVDF) membranes. PVDF membranes were then blocked with 5% solution of nonfat milk for 1 hour. Membranes were subsequently incubated with primary antibody against VMA21 (Origene; TA336068) or GAPDH (Origene; TA890003) according to the manufacturer's protocol. Then the membranes were washed three times and incubated with the appropriate amount of secondary antibodies for 1 hour at 37°C.
2.5 Cell counting kit-8 assay
Cells were seeded in 96-well plates at a density of 1 × 103 per well and cultured for 24, 48, 72 or 96 hours. Cell counting kit-8 (CCK-8) solution was added into 96-well plates after cell growth. Then, the cells were incubated for 4 more hours. After 4 hours, cell supernatant was removed and the optical density value was detected at 450 nm.
2.6 Colony formation assay
After 24 hours of transfection, A549 and H1299 cells were seeded into six-well plates at a density of 1 × 103 cells per well. The medium was replaced every 72 hours during growth. After 12 days of incubation, cells were fixed with methanol and stained with crystal violet. The colonies were counted when they contained more than 50 cells.
2.7 Luciferase reporter assay
All luciferase reporter vectors (ZFPM2-AS1-WT, ZFPM2-AS1-MUT, VMA21-WT, and VMA21-MUT) were constructed from empty pGL3 vectors (Promega, Madison, WI). A549 and H1299 cells were seeded into 24-well plate and grew until reaching 70% confluence. Cells were then cotransfected with luciferase reporter vectors and NC mimics, miR-18b-5p mimics or miR-18b-5p mimics + pcDNA3.1/ZFPM2-AS1 with Lipofectamine 2000. After 48 hours, luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions.
2.8 RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assay was carried out with EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) under the manufacturer's protocol. Then, A549 and H1299 cells were lysed with RNA lysis buffer. Cell lysis solutions were added with magnetic beads coated with anti-Ago2 antibodies (Millipore) or anti-IgG antibodies (Millipore). After 2 hours of incubation at 4°C, co-precipitated RNAs were extracted and examined by quantitative reverse transcription polymerase chain reaction (RT-qPCR).
2.9 RNA pull down
RNA pull-down was carried out to detect the binding capacity between miR-19b-5p and ZFPM2-AS1 or VMA21 in A549 and H1299 cells. ZFPM2-AS1-WT, ZFPM2-AS1-Mut, VMA21-WT, VMA21-Mut, and negative control (NC) were biotinylated and transfected into cells. Cell lysates were then added with streptavidin-coupled magnetic beads for 2 more hours at 37°C. Then, the RNA complex was eluted and miR-18b-5p was quantified through RT-qPCR.
2.10 Statistical analysis
Experiment data were expressed as mean ± SD based on results from three independent experiments. The Student t test and one-way analysis of variance were applied when comparing differences between groups. Survival curves were plotted by the Kaplan-Meier method. Data were considered as statistically significant when P value less than 0.05.
3 RESULTS
3.1 lncRNA ZFPM2-AS1 promotes proliferation in LUAD
Based on data collected from Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/), we found that the expression of lncRNA ZFPM2-AS1 was positively associated with the survival rate of LUAD patients (Figure 1A). Also, GEPIA showed that ZFPM2-AS1 was highly expressed in LUAD tissues than in normal tissues (Figure 1B). Furthermore, the expression of ZFPM2-AS1 was significantly higher in LUAD cells compared to HBE cells (Figure 1C). For experiment convenience, A549 and H1299 cells lines would be applied in the following research for their relatively high expression of ZFPM2-AS1.
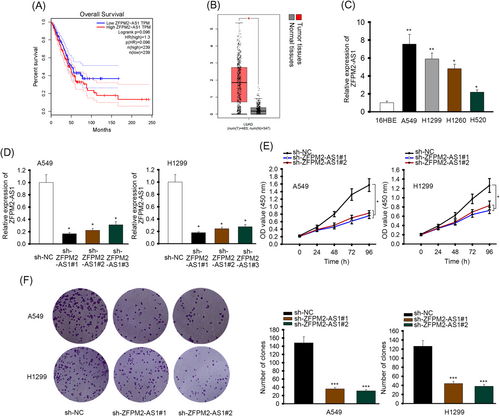
lncRNA ZFPM2-AS1 promoted proliferation in LUAD. A, Kaplein-Meier analysis of ZFPM2-AS1 in LUAD patients, data collected from GEPIA database (http://gepia.cancer-pku.cn). B, RT-qPCR of ZFPM2-AS1 in LUAD tissues and normal tissues, data collected from GEPIA database. C, RT-qPCR of ZFPM2-AS1 in LUAD cells (A549, H1299, H1260, and H520) and human bronchus epithelial cell (16HBE). D, RT-qPCR of ZFPM2-AS1 in cells transfected with sh-ZFPM2-AS1#1/2/3. E, CCK-8 assay in cells transfected with sh-ZFPM2-AS1. F, Colony formation assay in cells transfected with sh-ZFPM2-AS1. CCK-8, cell counting kit-8; GEPIA, Gene Expression Profiling Interactive Analysis; lncRNA, long noncoding RNA; LUAD, lung adenocarcinoma; RT-qPCR, quantitative reverse transcription polymerase chain reaction; ZFPM2-AS1, zinc finger protein multitype 2 antisense RNA 1. *P < 0.05, **P < 0.01, ***P < 0.001
So we performed assays to see if the expression of ZFPM2-AS1 would interfere proliferation of LUAD cells in return. The expression of ZFPM2-AS1 was effectively knocked down in cells transfected with sh-ZFPM2-AS1 (Figure 1D). Sh-ZFPM2-AS1#1 and sh-ZFPM2-AS1#2 would be applied in the following assays due to their relatively high knockdown efficiency. In CCK-8 assay, cells transfected with sh-ZFPM2-AS1 showed a weaker proliferation ability than the negative control group (Figure 1E). Also, cells transfected with sh-ZFPM2-AS1 formed much less colonies than cells transfected with sh-NC (Figure 1F). So based on these assays, lncRNA ZFPM2-AS1 promotes proliferation in LUAD.
3.2 ZFPM2-AS1 binds with and negatively regulates expression of miR-18b-5p
Since the function of ZFPM2-AS1 in LUAD had already been confirmed, we searched for its downstream gene to explore the concrete mechanism. Based on results predicted by starBase v3.0 (http://starbase.sysu.edu.cn/index.php), ZFPM2-AS1 has potential binding sites with 10 miRNAs. Among them, miR-18b-5p has been verified in most types of cancers based on results from Pan-Cancer analysis. So miR-18b-5p was selected for further detection.
We inspected the expression of miR-18b-5p and found its overall expression was lower in LUAD cells (Figure 2A). So we ran a series of test to verify the binding capacity between miR-18b-5p and ZFPM2-AS1. The potential binding site between them is listed in Figure 2B. Luciferase reporter assay showed that luciferase activity of vectors containing wild type ZFPM2-AS1 was lowered by miR-18b-5p mimics, while mutant ZFPM2-AS1 vectors showed no significant response to miR-18b-5p mimics (Figure 2C). Also, the majority of miR-18b-5p was detected from biotinylated wild type ZFPM2-AS1 group instead of biotinylated mutant ZFPM2-AS1 group in RNA pull-down (Figure 2D). Both assays indicated miR-18b-5p and ZFPM2-AS1 could bind at the predicted site. We also detected the influence of ZFPM2-AS1 on miR-18b-5p expression. The overexpression efficiency was verified before further experiments (Figure 2E). Figure 2F shows that overexpression of ZFPM2-AS1 would suppress the expression of miR-18b-5p. So based on these assays, ZFPM2-AS1 binds with and negatively regulates the expression of miR-18b-5p.
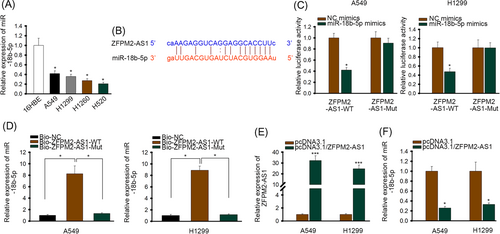
ZFPM2-AS1 bound with and negatively regulated expression of miR-18b-5p. A, RT-qPCR of miR-18b-5p in LUAD cells and human bronchus epithelial cells. B, The potential binding site between ZFPM2-AS1 and miR-18b-5p, predicted by starBase v3.0 (http://starbase.sysu.edu.cn/index.php). C, Luciferase reporter assay of wild type and mutant ZFPM2-AS1 vectors in cells transfected with NC mimics or miR-18b-5p. D, RNA pull down of miR-18b-5p in cells transfected with biotinylated wild type or mutant ZFPM2-AS1. E, RT-qPCR of ZFPM2-AS1 in cells transfected with pcDNA3.1 or pcDNA3.1/ZFPM2-AS1. F, RT-qPCR of miR-18b-5p in cells transfected with pcDNA3.1 or pcDNA3.1/ZFPM2-AS1. LUAD, lung adenocarcinoma; miR, microRNA; Mut, mutant; NC, negative control; RT-qPCR, quantitative reverse transcription polymerase chain reaction; WT, wild-type; ZFPM2-AS1, zinc finger protein multitype 2 antisense RNA 1. *P < 0.05, ***P < 0.001
3.3 miR-18b-5p binds with and negatively regulates the expression of VMA21
The target gene of miR-18b-5p was searched through PicTar database (https://pictar.mdc-berlin.de). Results showed that 47 mRNAs could bind with miR-18b-5p. Among them, VMA21 had been verified by most experiments at the number of 30. So VMA21 was selected for further detection.
The expression of VMA21 was inspected and we found that its expression in LUAD cells was significantly higher than in HBE cells (Figure 3A). So the binding capacity between VMA21 and miR-18b-5p was verified. The binding site was shown in Figure 3B. In luciferase reporter assay, the luciferase activity of vectors containing wild type VMA21 was suppressed by miR-18b-5p mimics, while mutant VMA21 vectors showed no significant response (Figure 3C). Also, the majority of miR-18b-5p enriched in biotinylated wild type VMA21 group instead of biotinylated mutant VMA21 group (Figure 3D). Furthermore, both mRNA and protein level of VMA21 was decreased by miR-18b-5p mimics (Figure 3E and 3F). So based on these assays, miR-18b-5p binds with and negatively regulates the expression of VMA21.
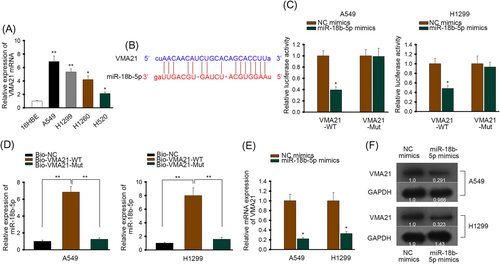
miR-18b-5p bound with and negatively regulated the expression of VMA21. A, RT-qPCR of VMA21 mRNA in LUAD cells and human bronchus epithelial cells. B, The potential binding site between VMA21 and miR-18b-5p, predicted by PicTar database (https://pictar.mdc-berlin.de). C, Luciferase reporter assay of wild type and mutant VMA21 vectors in cells transfected with NC mimics or miR-18b-5p mimics. D, RNA pull down of miR-18b-5p in cells transfected with biotinylated wild type or mutant VMA21. E, RT-qPCR of VMA21 mRNA in cells transfected with NC mimics or miR-18b-5p. F, Western blot of VMA21 in cells transfected with NC mimics or miR-18b-5p mimics. LUAD, lung adenocarcinoma; miR, microRNA; RNA, messenger RNA; Mut, mutant; NC, negative control; RT-qPCR, quantitative reverse transcription polymerase chain reaction; WT, wild-type. *P < 0.05, **P < 0.01
3.4 ZFPM2-AS1 regulates VMA21 expression by competitively binding with miR-18b-5p
Plenty of research have proved that lncRNAs might competitively bind with miRNAs, thus regulated the target gene expression of miRNAs.11-13 In these circumstances, miRNAs and Ago2 would form RISCs which could bind with both specific lncRNAs and mRNAs. In this way, the expression of mRNAs would be regulated by lncRNAs through miRNAs. Since we have confirmed the binding capacity between miR-18b-5p and ZFPM2-AS1 or VMA21, we would run a series of tests to see if ZFPM2-AS1 regulated VMA21 expression through such ceRNA pattern.
In Figure 4A, RIP assay demonstrated that the expression of miR-18b-5p, ZFPM2-AS1 and VMA21 could all be detected from Ago2 group. The RIP assay indicated that both ZFPM2-AS1 and VMA21 could bind with the RISC formed by miR-18b-5p and Ago2. Furthermore, both the mRNA and protein level of VMA21 could be promoted by overexpression of ZFPM2-AS1 (Figure 4B and 4C). Luciferase reporter assay was also performed to verify such ceRNA axis. In Figure 4D, the luciferase activity of wild type VMA21 was suppressed by overexpression of miR-18b-5p mimics, but was partly rescued by overexpression of ZFPM2-AS1. So based on these assays, ZFPM2-AS1 regulates VMA21 expression by competitively binding with miR-18b-5p.
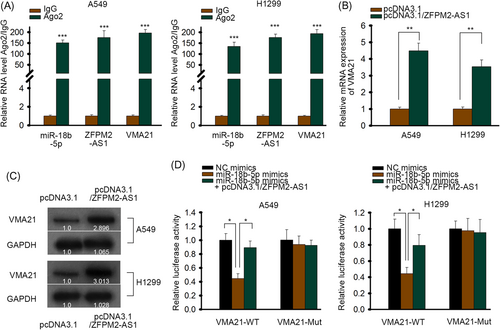
ZFPM2-AS1 regulated VMA21 expression by competitively binding with miR-18b-5p. A, RIP assay of miR-18b-5p, ZFPM2-AS1, and VMA21 in cells transfected with antibodies against IgG or Ago2. B, RT-qPCR of VMA21 mRNA in cells transfected with pcDNA3.1 or pcDNA3.1/ZFPM2-AS1. C, Western blot of VMA21 in cells transfected with pcDNA3.1 or pcDNA3.1/ZFPM2-AS1. D, Luciferase reporter assay of wild type and mutant VMA21 vectors in cells transfected with NC mimics, miR-18b-5p mimics or miR-18b-5p mimics + pcDNA3.1/ZFPM2-AS1. Ago2, Argonaute-2; IgG, immunoglobulin G; miR, microRNA; mRNA, messenger RNA; Mut, mutant; NC, negative control; RIP, RNA immunoprecipitation; RT-qPCR, quantitative reverse transcription polymerase chain reaction; WT, wild-type; ZFPM2-AS1, zinc finger protein multitype 2 antisense RNA 1. *P < 0.05, **P < 0.01, ***P < 0.001
3.5 ZFPM2-AS1 regulates cell proliferation via miR-18b-5p/VMA21 axis in LUAD
Since the ceRNA axis of ZFPM2-AS1/miR-18b-5p/VMA21 has already been confirmed, we ran a series of rescue assays to see if ZFPM2-AS1 regulated LUAD cell proliferation through this axis. The expression of VMA21 in LUAD cells was promoted for further experiments (Figure 5A). CCK-8 assays revealed that cell proliferation ability was weakened by knockdown of ZFPM2-AS1, but this trend was partly counteracted by overexpression of VMA21 (Figure 5B). Also, in colony formation assay, the number of colonies was decreased in sh-ZFPM2-AS1#1 group while the number in sh-ZFPM2-AS1#1 + pcDNA3.1/VMA21 group was partly restored (Figure 5C). So based on these assays, ZFPM2-AS1 regulates cell proliferation via miR-18b-5p/VMA21 axis in LUAD. The detailed scheme is presented in Figure 6.
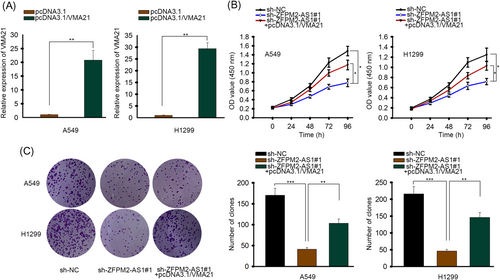
ZFPM2-AS1 regulated cell proliferation via miR-18b-5p/VMA21 axis in LUAD. A, RT-qPCR of VMA21 mRNA in cells transfected with pcDNA3.1 or pcDNA3.1/VMA21. B, CCK-8 assay in cells transfected with sh-NC, sh-ZFPM2-AS1#1 or sh-ZFPM2-AS1#1 + pcDNA3.1/VMA21. C, Colony formation assay in cells transfected with sh-NC, sh-ZFPM2-AS1#1 or sh-ZFPM2-AS1#1 + pcDNA3.1/VMA21. CCK-8, cell counting kit-8; LUAD, lung adenocarcinoma; miR, microRNA; mRNA, messenger RNA; NC, negative control; RT-qPCR, quantitative reverse transcription polymerase chain reaction; ZFPM2-AS1, zinc finger protein multitype 2 antisense RNA 1. *P < 0.05, **P < 0.01, ***P < 0.001
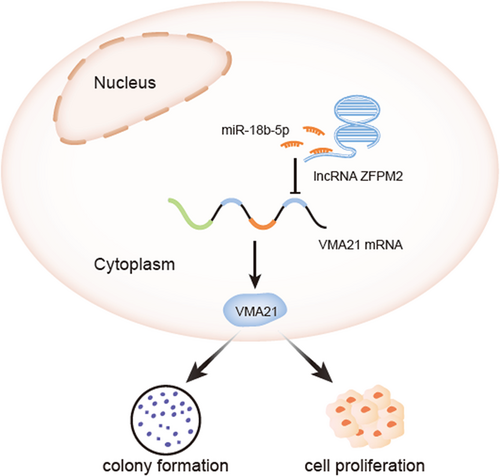
Scheme illustrating the mediation procedure of ZFPM2-AS1. lncRNA, long noncoding RNA; miR, microRNA; ZFPM2-AS1, zinc finger protein multitype 2 antisense RNA 1
4 DISCUSSION
Although great progress has been achieved in cancer treatment and overall medical technology, the treatment in LUAD is still not optimistic. The 5-year survival rate of lung cancer patients is still depressing.14 Many factors may lead to LUAD, including smoking, aging, and air pollution. Therefore, thorough detection of tumorigenesis and cancer progression is badly called for. Fortunately, the field of gene interference has witnessed inspiring progress. Research have proved that the abnormal expression of lncRNAs might be an important inducing factor of various cancers. For example, LINC00467 was proved to promote tumor cell survival.15 Or, UICLM promotes liver metastasis of colorectal cancer.16 Also, LUCAT1 was found to promote tumorigenesis of colorectal cancer.17 However, due to their massive amount and complicated mechanism, many lncRNAs have not been thoroughly detected. ZFPM2-AS1 is one of them. In open research of recent years, ZFPM2-AS1 were only demonstrated to promote gastric carcinogenesis.10 In this study, we found that high expression of ZFPM2-AS1 is associated with poor prognosis in LUAD. So the function and molecule mechanism of ZFPM2-AS1 in LUAD is detected by a series of studies in this study.
lncRNAs were found to mediate cancer progression in various ways. One of these ways is the ceRNA mechanism. Since being brought up by Pier Pablo Pandolfi in 2011, the ceRNA mechanism has become an important tool in cancer research. CeRNA hypothesis pictured a clear relationship between lncRNA, miRNA, and mRNAs. To be specific, the ceRNA hypothesis believes that miRNA and Ago2 forms RISCs which dissolves the target gene of miRNA. However, some specific lncRNAs can competitively bind with RISCs to free mRNAs from being dissolved. In this way, lncRNAs indirectly regulate the expression of mRNAs, thus regulating cancer progression. For example, LINC02532 was found to promote gastric cancer progression via sponging miR-129-5p and miR-490-5p.18 Or, UCA1 acts as a ceRNA to regulate Slug in glioma.19 Also, SNHG6 has been proved to promote colorectal progression via miR-760/FOXC1 axis.20 In this study, miR-18b-5p could bind with both ZFPM2-AS1 and VMA21. In the meantime, ZFPM2-AS1 negatively mediated miR-18b-5p expression and miR-18b-5p negatively mediated VMA21 expression. Furthermore, both ZFPM2-AS1 and VMA21 were found to adjoin to the RISC constructed by miR-18b-5p and Ago2. ZFPM2-AS1 regulated VMA21 expression through miR-18b-5p. Thereby the ceRNA pattern was confirmed. Through a series of rescue assays, ZFPM2-AS1 was demonstrated to mediate LUAD cell proliferation through competitively binding with miR-18b-5p to regulate VMA21 expression. To sum up, lncRNA ZFPM2-AS1 promotes proliferation via miR-18b-5p/VMA21 axis in lung adenocarcinoma.
For the first time, the function and molecule mechanism of ZFPM2-AS1 were studied in LUAD by this study. The ZFPM2-AS1/miR-18b-5p/VMA21 axis is brand new. ZFPM2-AS1 might serve as a novel target in cancer research.
ACKNOWLEDGMENT
The authors thank all those involved in this study. The study was supported by the Shanghai Minhang District Central Hospital Youth Research Fund (2018MHLC01).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
MX participated in the design of this study. WT and SY contributed significantly to manuscript preparation. ZY and QP performed the experimental data record and analysis. FJ and XG wrote the manuscript. All authors performed their constructive discussions in this study.