Overexpression of miR-623 suppresses progression of hepatocellular carcinoma via regulating the PI3K/Akt signaling pathway by targeting XRCC5
Abstract
It has been reported that miR-623 is deregulated in lung adenocarcinoma and inhibits tumor growth and invasion. However, it is unclear whether miR-623 has a role in the progression of hepatocellular carcinoma (HCC). Herein, we found that miR-623 was significantly downregulated in HCC, and that its expression was related to poor clinical outcomes of patients with HCC. Upregulation of miR-623 decreased cell proliferation, viability, migration, and invasion and further promoted apoptosis in 7721, Huh7, and Bel-7402 cells. Moreover, we also observed that miR-623 regulated the phosphoinositide 3-kinase/protein kinase B (PI3K/Akt), Wnt/β-catenin, and extracellular regulated protein kinases/c-Jun N-terminal kinase (ERK/JNK) signaling pathways as well as the expression level of related proteins. Further, X-ray repair cross complementing 5 (XRCC5) was a direct target for miR-623, and the suppression of PI3K/Akt, Wnt/β-catenin, and ERK/JNK signaling pathways and cell proliferation and invasion abilities caused by miR-623 in HCC cells was significantly reversed by the upregulation of XRCC5. Collectively, our data suggested that miR-623 suppressed the progression of HCC by regulating the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways by targeting XRCC5 in HCC in vitro, indicating that miR-623 may be a target for the therapy of HCC.
1 INTRODUCTION
It is estimated that 600 000 people die of hepatocellular carcinoma (HCC) worldwide each year, with the majority of patients found in Asia and Africa, and the rate of incidence is still rising.1, 2 It is imperative to explore the physiological and molecular mechanisms of HCC to find potential therapeutic targets that establish new treatment programs.
MicroRNAs (miRNAs) can bind to their target gene's 3′-untranslated region (UTR) to regulate their expression.3 Increasing evidence shows that miRNAs have the potential to regulate cell proliferation, apoptosis, and invasion by either inhibiting target gene translation or promoting degradation of the messenger RNA (mRNA), thereby affecting tumor progression and metastasis.4, 5 In addition, it has been reported recently that the majority of chronic lymphoblastic leukemia carry deletions of miR-15a and miR-16-1 which have multiple targets, and the monoclonal antibodies against their target genes can kill chronic lymphoblastic leukemia cells, such as venetoclax against Bcl-2 and monoclonal antibodies against ROR1, providing a new potential strategy for cancer treatment.6 Studies have confirmed that some miRNAs were deregulated in HCC and that they were associated with HCC progression and prognosis of patients.7-12 miR-623, a newly identified miRNA, has recently been reported to be downregulated in gastric cancer tissues and cell lines, and it's upregulation inhibits the growth of gastric cancer.13 However, it is unclear whether miR-623 has a role in the progression of HCC.
X-ray repair cross complementing 5 (XRCC5) encodes a heterodimer of Ku (Ku80), which has the potential to activate DNA-dependent protein kinase. Further studies have shown that XRCC5 functions in the cellular process, primarily by maintaining telomeres, gene transcription, and the regulation of apoptosis and G2/M phase arrest.14-16 Recently, XRCC5 was reported to be involved in the progression of cancers, with the upregulation of XRCC5 being observed in cancer tissues, including bladder cancer, non–small cell lung cancer , and gastric cancer.17-20 Silencing the expression of XRCC5 could thus significantly inhibit tumor growth.21 Therefore, targeting XRCC5 might be a potential method to inhibit tumor growth.
As is commonly known, the phosphoinositide 3-kinase/protein kinase B (PI3K/Akt), Wnt/β-catenin, and extracellular regulated protein kinases/c-Jun N-terminal kinase (ERK/JNK) pathways are both critical for cell proliferation, apoptosis, and differentiation and have been confirmed to be involved in HCC progression.22-25 Inhibiting these signaling pathways is considered to be an effective method for cancer therapy.
We have therefore investigated the role and underlying mechanism of miR-623 in tumor progression of HCC. Our study is the first to confirm that miR-623 regulated the growth, invasion, and survival of HCC cells through targeting XRCC5.
2 MATERIALS AND METHODS
2.1 Patients and specimens
The study was approved by the Research Ethics Committee of Ningbo No.2 Hospital. A total of 53 pairs of human HCC tissues and their corresponding adjacent liver tissues were collected from patients (male/female: 38/15, age ≤50/>50 years: 29/24) undergoing liver resection surgery at the Ningbo No.2 Hospital between February 2012 and January 2018. Written informed consent was obtained from all the patients. Clinicopathologic histories for these patients are shown in Table 1. Patients were subcategorized in either the miR-623 downregulated group (n = 39) or the intact group (n = 14). We did this by comparing the median value of miR-623 expression among the HCC tissue samples that were analyzed.
| Variables | Total cases (n = 53) | miR-623 expression level | P value | |
|---|---|---|---|---|
| Downregulated (n = 9) | Intact (n = 14) | |||
| Age, y | .4402 | |||
| ≤50 | 29 | 20 | 9 | |
| >50 | 24 | 19 | 5 | |
| Sex | .473 | |||
| Male | 38 | 29 | 9 | |
| Female | 15 | 10 | 5 | |
| Liver cirrhosis | .104 | |||
| None/mild | 14 | 8 | 6 | |
| Moderate/severe | 39 | 31 | 8 | |
| HBV-DNA (copies/mL) | .010a | |||
| ≤5 × 102 | 16 | 8 | 8 | |
| >5 × 102 | 37 | 34 | 6 | |
| HCV | .257 | |||
| Positive | 13 | 8 | 5 | |
| Negative | 40 | 31 | 9 | |
| Tumor multiplicity | .314 | |||
| Solitary | 36 | 28 | 8 | |
| Multiple | 17 | 11 | 6 | |
| Tumor diameter, cm | .0278a | |||
| ≤5 | 21 | 12 | 9 | |
| >5 | 32 | 27 | 5 | |
| AFP, ng/mL | .872 | |||
| <25 | 18 | 13 | 5 | |
| ≥25 | 35 | 26 | 9 | |
| Vascular invasion | .0136a | |||
| Yes | 23 | 13 | 10 | |
| No | 30 | 26 | 4 | |
| Child-Pugh score | .645 | |||
| A | 33 | 25 | 8 | |
| B | 20 | 14 | 6 | |
| Stage (UICC) | .0328a | |||
| I and II | 18 | 10 | 8 | |
| III and IV | 35 | 29 | 6 | |
- Abbreviations: AFP, alpha-fetoprotein; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; UICC, Union for International Cancer Control.
- a Statistically significant (P < .05).
2.2 Cells culture
Liver cancer cell lines Hep3B, 7721, Huh7, HepG2, and Bel-7402 cells were seeded in Roswell Park Memorial Institute-1640 (HyClone) medium enriched with 10% fetal bovine serum (FBS). 7721, Huh7, and Bel-7402 cells were transiently transfected with either the pCMV-MIR-miR-623 plasmid (2.5 µg) when they grew to 80% fusion, and pCMV-MIR was performed as negative control (NC).
For XRCC5 transfection, cells (2 × 104) were grown in a six-well plate and cultured for 24 hours and transfected with siRNA-XRCC5 (50 nM; Ribobio) using Lipofectamine 2000 to block the expression of XRCC5, and scrambled siRNA was used as negative control. The expressing vector (pcDNA3.1-XRCC5; Ribobio) was conducted and cells were cotransfected with miR-623 and XRCC5 using Lipofectamine 2000, and the blank vector was used as negative control. The siRNA-XRCC5 sequence was 5′-GGATGTGATTCAACATGAA-3′.
2.3 Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA from HCC cells were extracted by means of the Ultrapure RNA Kit (Cat #CW057; CWBIO). Complementary DNA was reverse-transcribed from RNA by use of the miRNA cDNA Synthesis Kit (Cat #CW2141; CWBIO). The related expression of miR-623 was then measured by the miRNA qPCR Assay Kit (Cat #CW2142S; CWBIO). The primer used in this study was synthesized from (Ribobio) miR-623: 5′-AUCCCUUGCAGGGGCUGU-3′; XRCC5: 5′-CCATGAGCTTGGCAAAGAAAG-3′, 5′-GTGCAGCAGACACTGAAATAATC-3′; β-actin: 5′-CCCGAGCCGTGTTTCCT-3′, 5′-GTCCCAGTTGGTGACGATGC-3′.
2.4 Western blot assay
After being transfected for 48 hours, 20 μg of samples extracted from cells were then loaded to each lane in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and were electrotransferred to the polyvinylidene fluoride membrane, which was then blocked with 5% nonfat milk followed by incubation with primary antibodies and with secondary antibodies for 1 hour. The ECL substrate was applied to visualize the protein bands. Glyceraldehyde 3-phosphate dehydrogenase was used as the loading control and the relative expression of proteins was normalized to NC group.
2.5 CCK-8 assay
After being transfected for 24 hours, cells were grown in a 96-well plate and then incubated with cell counting kit-8 (CCK-8) reagent (10 μL/well; CWBIO) for 1.5 hour. The optical density value was measured every 24 hours at 450 nm.
2.6 Colony formation assay
Cells were grown into a 6-cm plate and incubated with 5% CO2 for 10 days. After washing with phosphate-buffered saline, colonies were fixed with 4% paraformaldehyde followed by staining with 500 μL of GIEMSA (Cat #AR-0752; DingGuo Bio, Shanghai, China) for 10 minutes.
2.7 Transwell assay
Transwell chambers coated with Matrigel were performed for assessment of cell invasion. About 1 × 104 of HCC cells were seeded in the upper compartment, and the lower compartment was filled with 600 µL of medium supplemented with FBS. After incubation for 24 hours, the invaded cells were treated with 4% paraformaldehyde, with 0.1% crystal violet and then stained for 5 minutes. The stained cells were captured (magnification, ×100). Transwell chamber did not require Matrigel in the migration assay.
2.8 Apoptosis assay
The Annexin V-FITC- PI Apoptosis Detection Kit (4A Biotech, Beijing, China) was used to measure the rate of apoptosis in HCC cells. The rate of apoptosis was calculated by using the BD FACSDiva Software (BD FACSC anto II, BD Biosciences, Bedford MA).
2.9 Dual-luciferase reporter assay
A total of 5 × 105 HCC cells were inoculated into culture plates. After incubation for 24 hours, luciferase reporter plasmids, pmirGLO-XRCC5-3′UTR-wild type (wt) or pmirGLO- XRCC5-3′UTR-mutant type (mut), were cotransfected with either pCMV-pMIR-miR-623 (miR-623) or pCMV-MIR (NC) into the HCC cells. The luciferase activity of cells was measured when cells were cultured for 48 hours.
2.10 Statistical analysis
The data were shown as means ± SD and statistically analyzed by the SPAA 18.0 saftware (SPSS Inc., Chicago, IL). The differences between groups were analyzed using the Student t test or one-way analysis of variance. The association between two variables was assessed by the χ2 test. Pearson's correlation coefficient was used for correlation analysis.
3 RESULTS
3.1 Downregulation of miR-623 is correlated with poor clinical outcomes of HCC
Our data illustrated that miR-623 was downregulated in 73.6% (39/53) HCC tissues (P < .01; Figure 1A) when compared with the corresponding normal liver tissues. Moreover, the expression of miR-623 was correlated with HBV-DNA load (P = .010), tumor size (P = .0278), vascular invasion (P = .0136), and tumor stages (P = .0328) (Table 1). Collectively, miR-623 may play a role in the progression of HCC.
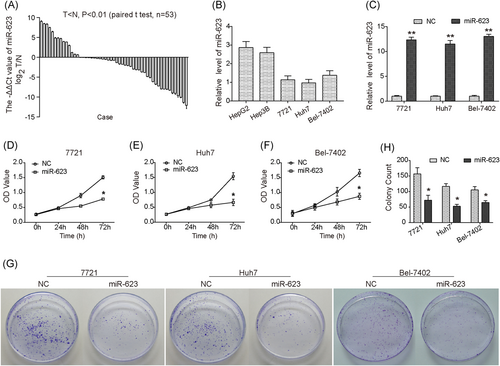
miR-623 is downregulated in HCC tissues and inhibits cell proliferation and viability of HCC cells. (A) Downregulation of miR-623 in HCC tissues (n = 53) compared with adjacent liver tissues (P < .01; n = 53). (B) Relative expression level of miR-623 in different liver cancer cell lines. (C) Relative expression of miR-623 in three HCC cell lines transfected with miR-623 or miR-NC. (D-F) CCK-8 assay showed that overexpression of miR-623 inhibited proliferation of 7721 (D), Huh7 (E), and Bel-7402 (F) cells. (G) Colony formation assay showed that overexpression of miR-623 inhibited cell growth and viability of 7721, Huh7, and Bel-7402 cells in vitro. (H) Quantitative analysis of colony formation assay. *P < .05, **P < .01 compared with negative control (NC). CCK-8, Cell Counting Kit-8; HCC, hepatocellular carcinoma
3.2 Overexpression of miR-623 suppresses the proliferation and viability of HCC cells
The expression of miR-623 in different liver cancer cell lines were detected, and we observed that the level of miR-623 in 7721, Huh7, and Bel-7402 cells was much low (Figure 1B; P < .0001). The pCMV-MIR-miR-623 plasmid was transfected into 7721, Huh7, and Bel-7402 cells to investigate the effects of miR-623 on HCC cells (Figure 1C). As shown in Figure 1 D, overexpression of miR-623 caused a significant reduction in the viability of 7721 cells. Similar inhibition caused by upregulation of miR-623 was also observed in Huh7 and Bel-7402 cells (Figure 1E, F). Moreover, our results showed that the colony count was markedly decreased in miR-623 overexpressed cells (Figure 1G,H). Collectively, these results suggested an antigrowth role of miR-623 in HCC cells.
3.3 Upregulation of miR-623 inhibits cell migration and invasion in HCC cells in vitro
To investigate the effect of miR-623 on HCC cell mobility, a Transwell assay was performed. Our data confirmed that upregulation of miR-623 reduced the migration abilities of 7721, Huh7, and Bel-7402 cells (Figure 2A-D). In addition, cell invasion abilities were obviously suppressed by upregulation of miR-623 (Figure 2E-H).
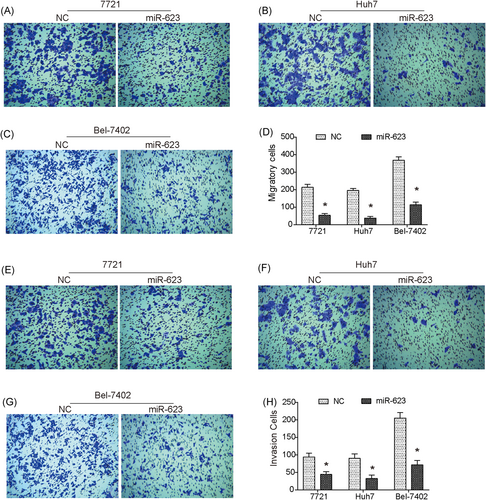
Overexpression of miR-623 inhibits migration and invasion of HCC cells in vitro. (A-C) Transwell assay showed that overexpression of miR-623 significantly suppressed migration of 7721 (A), Huh7 (B), and Bel-7402 (C) cells. (D) Quantitative analysis of Transwell migration assay. (E-G) Transwell invasion assay revealed that the invasion ability of 7721 (E), Huh7 (F), and Bel-7402 (G) cells was significantly decreased by upregulation of miR-623. (H) Quantitative analysis of Transwell invasion assay. *P < .05 compared with negative control (NC). HCC, hepatocellular carcinoma
3.4 Overexpression of miR-623 induces apoptosis
We found that rate of cell apoptosis was significantly promoted by upregulation of miR-623 in HCC cells (Figure 3A,B). Moreover, we observed that the expression of Bcl-2 was downregulated in miR-623 overexpressed cells, and the level of both total casapse3 and total caspase9 was decreased, while the expression of Bax, cleaved caspase3 and cleaved caspase9 was significantly increased (Figure 3C,D). These data suggested that upregulation of miR-623 induced apoptosis in HCC cells might be done through the mitochondrial apoptotic pathway.
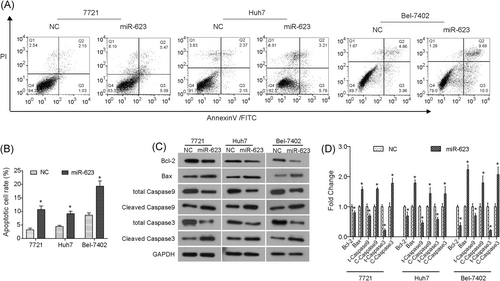
Upregulation of miR-623 induces apoptosis of HCC cells in vitro. A,B, Overexpression of miR-623 induced the rate of apoptosis in HCC cells. C,D, Western blot analysis results revealed that miR-623 impacted expression of apoptosis-related proteins, Bcl-2, Bax, total caspase9 (t-caspase9), cleaved caspase9 (c-caspase9), total caspase3 (t-caspase3), and cleaved caspase3 (c-caspase3). *P < .05 compared with negative control (NC). GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCC, hepatocellular carcinoma
3.5 Upregulation of miR-623 suppresses the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways
The PI3K/Akt pathway is indispensable in cell growth and survival as it regulates cell cycle–related proteins and Bcl-2-related proteins, both of which have been confirmed to be involved in HCC progression.22, 25 As shown in Figure 4A,B, we found that overexpression of miR-623 did not affect the total level of Akt and mammalian target of rapamycin (mTOR), whereas phosphorylated form p-Akt and p-mTOR was significantly inhibited in HCC cells. In addition, overexpression of miR-623 also downregulated the expressions of cyclin D1 and p70S6K in HCC cells (Figure 4A,B). Collectively, our data suggested that upregulation of miR-623 inhibited the PI3K/Akt signaling pathway.
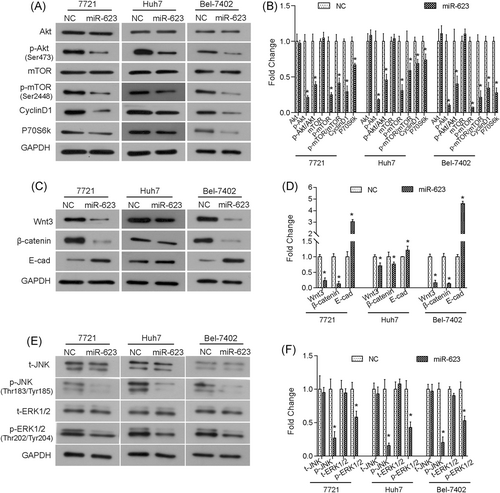
Overexpression of miR-623 downregulates the activation of PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways. A,B, Overexpression of miR-623 inhibited the expression of PI3K/Akt pathway-related proteins in HCC cells. C,D, Overexpression of miR-623 inhibited the expression of Wnt/β-catenin pathway-related proteins. E,F, Western blot analysis showed that upregulation of miR-623 suppressed the activation of the ERK/JNK pathway in HCC cells. t-JNK, total JNK; p-JNK, phosphorylated JNK; t-ERK1/2, total ERK1/2; p-ERK1/2, phosphorylated ERK1/2. *P < .05 compared with negative control (NC). Akt, protein kinase B; ERK, extracellular regulated protein kinases; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCC, hepatocellular carcinoma; JNK, c-Jun N-terminal kinase; mTOR, mammalian target of rapamycin; PI3K, phosphoinositide 3-kinase
Moreover, we found that the expression of Wnt3 and β-catenin were decreased in miR-623 overexpressed cells (Figure 4C,D). In addition, we observed that the expression levels of downstream protein E-cad were different in the three cell lines, which was due to the diversity of cell lines. However, the expression of E-cad was upregulated by miR-623 in 7721, Huh7, and Bel-7402 cells (Figure 4C,D). Together, these data suggested that the Wnt/β-catenin pathway was also inhibited by the overexpression of miR-623 in HCC cells in vitro. In addition, upregulation of miR-623 decreased the phosphorylated forms of ERK1/2 and JNK (Figure 4E,F), suggesting that upregulation of miR-623 could inhibit the ERK/JNK pathway in HCC cells.
3.6 miR-623 directly targets XRCC5 3ʹUTR in HCC
As shown in Figure 5A, we found that XRCC5 3′UTR had a putative binding site for miR-623 from the TargetScan database. To validate it, cells were cotransfected with miR-623 and pmirGLO-XRCC5 3′UTR-wt or pmirGLO-XRCC5 3′UTR-mut. We observed that miR-623 clearly inhibited the luciferase activity of XRCC5-3′UTR in HCC cells (Figure 5B), whereas the mutant counterpart blocked the same decrease caused by miR-623, indicating that XRCC5 was a target of miR-623. Western blot analysis revealed that the protein expression of XRCC5 was significantly downregulated by miR-623 (Figure 5C). Collectively, these results indicated that miR-623 directly bound to XRCC5 and downregulated its expression in HCC cells.
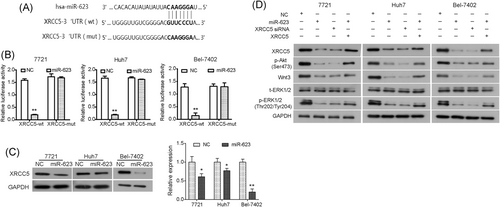
miR-623 directly targets XRCC5 in HCC cells in vitro. A, Predicted consequential pairing of XRCC5 mRNA and miR-623. B, Dual-luciferase report assays showed that miR-623 targeted XRCC5 in 7721, Huh7, and Bel-7402 cells. The pmirGLO-XRCC5-3′UTR (wt) or pmirGLO-XRCC5-3′UTR (mut) vectors were cotransfected into HCC cells with miR-623. C, Western blot analysis results showed that miR-623 downregulated expression of XRCC5 protein in 7721, Huh7, and Bel-7402 cells. D, HCC cells were transfected with negative control RNA, miR-623 vector or XRCC5 siRNA, and cells were cotransfected with miR-623 vector and XRCC5 vector. The expression of XRCC5, p-Akt, Wnt3, t-ERK1/2, and p-ERK1/2 was analyzed by Western blot. *P < .05, **P < .01 compared with negative control. Akt, protein kinase B; ERK, extracellular regulated protein kinases; HCC, hepatocellular carcinoma; mRNA, messenger RNA; mut, mutant type; NC, negative control; UTR, untranslated region; wt, wild type; XRCC5, X-ray repair cross complementing 5
3.7 Upregulation of XRCC5 reversed the miR-623-induced suppression for HCC
We predicted that the suppressive effect of miR-623 on growth and metastasis of HCC might be mediated by regulating XRCC5. To validate this prediction, HCC cells were cotransfected with XRCC5 over-expressing vector (lacking its 3′UTR) and pCMV-MIR-miR-623; and siRNA targeting XRCC5 was transfected into HCC cells to block XRCC5 (Figure S1A and S1B). As shown in Figure 5D, silencing XRCC5 significantly inhibited the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways by downregulating p-Akt, Wnt3, and p-ERK1/2, similar to that seen in HCC cells transfected with miR-623. Moreover, the inhibitory effect on p-Akt, Wnt3, and p-ERK1/2 caused by overexpression of miR-623 was significantly reversed by upregulation of XRCC5 (Figure 5D). Furthermore, as shown in Figure 6A-C, upregulation of XRCC5 (Figure S1C and S1D) reversed the suppression of cell proliferation induced by overexpression of miR-623 in HCC cells. In addition, the decreased migration and invasion capabilities induced by overexpression of miR-623 in HCC cells were significantly restored by the upregulation of XRCC5 (Figure 6D,E). Therefore, miR-623 may inhibit cell growth and invasion abilities of HCC cells through targeting XRCC5, and the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways may be involved.
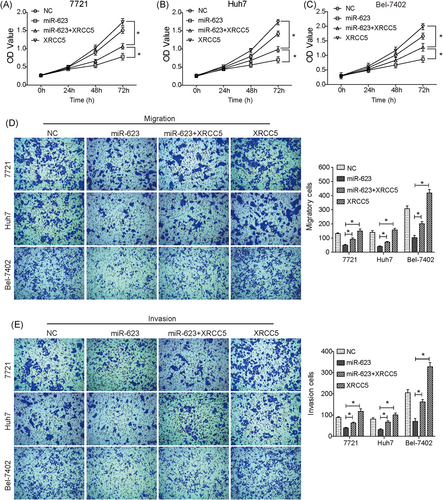
Upregulation of XRCC5 reverses the miR-623-induced suppression for HCC. (A-C) CCK-8 assay showed that the suppression on cell proliferation caused by overexpression of miR-623 in 7721 (A), Huh7 (B) and, Bel-7402 (C) cells was significantly reversed by the upregulation of XRCC5. (D) and (E) Upregulation of XRCC5 reversed the miR-623-induced suppression on migration (D) and invasion (E) capabilities in HCC cells. miR-623+XRCC5, cells cotransfected with pCMV-MIR-miR-623 plasmid and XRCC5 over-expressing vector; XRCC5, cells transfected with XRCC5 over-expressing vector. *P < .05. HCC, hepatocellular carcinoma; NC, negative control; XRCC5, X-ray repair cross complementing 5
4 DISCUSSION
Several different miRNAs have been found to function in the progression and metastasis in HCC.7-12 Jiang et al13 had revealed that miR-623 was downregulated in gastric cancer, and they further found that upregulation of miR-623 inhibited the proliferation in SGC-7901 and BGC-823 cells and enhanced the chemosensitivity of cells to 5-Fluorouracil. Herein, our data revealed that miR-623 was frequently downregulated in HCC tissues, and that it was significantly correlated with liver cirrhosis, HBV-DNA load, tumor size, vascular invasion, and tumor stages in patients with HCC. There are some biological differences among different hepatocellular carcinomas cell lines, hepatocellular carcinoma cell lines Bel-7402, Huh7, and 7721 were performed in this study. Bel-7402 cell line is alpha-fetoprotein (AFP)-positive and can form tumor nodules in treated Wistar rats; 7721 cell line is AFP positive, and its success rate of xenotransplantation is 100%; Huh7 cell line is HBV-negative and produces some cytoplasmic proteins, such as albumin, a-antitryptase, and AFP. Our results showed that upregulation of miR-623 reduced cell proliferation, viability, migration, and invasion of Bel-7402, Huh7, and 7721 cells. Furthermore, we found that miR-623 could induce apoptosis in a mitochondrial pathway by regulating the Bcl-2/Bax axis and caspase cascade. These results together suggested that miR-623 served as a tumor suppressor in the progression of HCC. We then elucidated the mechanism of miR-623 that underlying the inhibition on HCC tumor progression in vitro. Our data showed that miR-623 significantly inhibited the PI3K/Akt pathway; this was confirmed by the downstream proteins, cyclin D1, and P70S6k having downregulated expression. Cyclin D1 is a key regulator in cell cycle and has been reported to be upregulated in HCC and thought to be a potential target for HCC.26-28 It has also been demonstrated that miR-623 targeted cyclin D1 in gastric cancer cells.13 In this study, we found that miR-623 could inhibit the cyclin D1 expression in HCC cells. In addition, our data showed that miR-623 displayed a significant inhibition on the Wnt/β-catenin pathway in HCC cells and increasing the expression of the downstream protein E-cad. Furthermore, upregulation of miR-623 suppressed the activation of ERK/JNK pathway. Therefore, these results suggested that the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways were involved in the antitumor function of miR-623.
miRNAs function either by affecting the translation or promoting the degradation of target genes’ mRNA. Finding the target gene of miR-623 was therefore crucial to our study. In this study, our data revealed that miR-623 had the potential to reduce the expression of XRCC5, confirming that XRCC5 was a target of miR-623 in HCC cells. Moreover, the suppression in the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways caused by miR-623 was reversed by upregulation of XRCC5. Furthermore, the decreased growth, migration, and invasion abilities caused by overexpression of miR-623 were significantly reversed by the upregulation of XRCC5. Therefore, miR-623 inhibited the growth, migration, and invasion abilities of HCC cells through targeting XRCC5 by inactivating the PI3K/Akt, Wnt/β-catenin, and ERK/JNK pathways.
5 CONCLUSION
In conclusion, miR-623 was revealed to have the potential to inhibit the progression and metastasis of HCC in vitro through targeting XRCC5, indicating that miR-623 is a new therapeutic target for cancer. And the effects of miRNA-623 and XRCC5 on the progression of HCC tumor in vivo will be investigated in our future study.
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interests.