miR-4319 inhibited the development of thyroid cancer by modulating FUS-stabilized SMURF1
Abstract
Thyroid cancer, a common type of endocrine system cancer, has witnessed rising incidence and deaths over the past few years. The role of microRNAs is being increasingly discovered in a variety of cancers, including thyroid cancer. miR-4319 has been elucidated in several studies to exert an antitumor function in multiple cancers but has never been explored in thyroid cancer. Our study proposed to explore the function and modulatory mechanism of miR-4319 in thyroid cancer. First, we confirmed the downregulation of miR-4319 in thyroid cancer tissues and cells, and revealed the correlation of miR-4319 expression and clinical features in thyroid cancer. Functional assays illustrated that miR-4319 attenuated proliferation, migration, and epithelial-to-mesenchymal transition (EMT) in thyroid cancer. Mechanistically, we identified through the miRDB database and proved that miR-4319 targeted SMAD specific E3 ubiquitin protein ligase 1 (SMURF1). Furthermore, we discovered that miR-4319 competed with fused in sarcoma (FUS) to bind to SMURF1, inhibiting the stabilization of SMURF1 messenger RNA. Rescue assays suggested that miR-4319 retarded proliferation, migration, and EMT through SMURF1. Collectively, the results of this study showed that miR-4319 inhibited the development of thyroid cancer by modulating FUS-stabilized SMURF1, indicating miR-4319 as a potent biological target for thyroid cancer.
1 INTRODUCTION
Thyroid cancer is a prevalent malignancy of the human endocrine system.1 Past decades have witnessed a rising death rate due to the occurrence of thyroid cancer.2, 3 The canceration of thyroid cells involves a diversity of factors, such as the activation and inactivation of genes, methylation and mutation of DNA, and stimulation of the intracellular pathways.4, 5 Therefore, effective therapeutic biological targets are in imminent need to be identified in thyroid cancer.
MicroRNAs (miRNAs), classified as small noncoding RNAs with approximately 22 nucleotides, are endogenous molecules lacking in protein-coding potentials.6 Through complementary binding to 3' untranslated regions (3′UTR), miRNAs exhibit suppressive impacts on gene expression.7 Nowadays, miRNAs have gained a reputation as crucial factors in diverse biological activities, such as carcinogenesis, cell apoptosis, and proliferation.6 The implication of miRNAs in cancers has been increasingly documented,8, 9 including in thyroid cancer.10 miR-4319 has been recently illustrated to inhibit breast cancer and acute myeloid leukemia,11, 12 but its function and mechanism in thyroid cancer have never been investigated.
Interestingly, recent studies illustrated that miRNAs could either coordinate or compete with RNA binding proteins (RBPs) to regulate their shared target messenger RNAs (mRNAs).13, 14 Fused in sarcoma (FUS) is an RBP with multiple functions, including transcription activation in nucleus and mRNA stabilization in cytoplasm.15, 16 Also, FUS has been shown to play a part in thyroid cancer.17 However, its coordination or competition with miR-4319 in thyroid cancer has never been studied.
Smad ubiquitin regulatory factor 1 (SMURF1) is a C2-WW-HECT ubiquitin ligase. It is implicated in diverse biological processes, including viral autophagy embryogenesis and bone homeostasis.18-20 SMURF1 has been illustrated to promote proliferation, migration, and epithelial-to-mesenchymal transition (EMT) in diverse cancers21, 22 and has also been recently discovered to promote thyroid cancer progression.23 However, the interaction of SMURF1 with miR-4319 and FUS in thyroid cancer has never been revealed before.
The current study proposed to explore the role of miR-4319 in thyroid cancer and its modulatory mechanism on SMURF1.
2 MATERIALS AND METHODS
2.1 Tissue specimens
Fifty-six pairs of thyroid cancer tissues and matched paracancerous tissues were collected from patients who had received surgical dissection at the Xintai City People's Hospital of Shandong Province. The tissue samples were stored immediately in liquid nitrogen at −80℃ for later use. None of these patients had undergone any radiotherapy or chemotherapy before surgery.
2.2 Cell culture and reagents
Human normal thyroid epithelium cell line (Nthy-ori 3-1) and thyroid cancer cell lines (TPC, BHP5-16, BHP2-7, and K1) were offered by the tumor cell bank of The Chinese Academy of Sciences (Beijing, China). For cell incubation, 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA) was used to supplement the Roswell Park Memorial Institute-1640 medium (Invitrogen) with the addition of 100 U/mL penicillin as well as 100 mg/mL streptomycin (Invitrogen). A humid incubation atmosphere containing 5% CO2 at 37℃ was required.
2.3 Cell transfection
The oligo miR-4319 mimic and the negative controls (NC mimic) were produced by RiboBio Company (Guangzhou, China). The overexpression of FUS or SMURF1 was achieved by the transfection of pcDNA3.1 vectors integrated with the full sequences of FUS or SMURF1 (pcDNA3.1/FUS or pcDNA3.1/SMURF1). The silencing of FUS was realized using specific short hairpin RNAs (shRNAs) targeting FUS, with shNC acting as negative control. The plasmids were introduced into the thyroid cancer cells with the assistance of Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA). After 48 hours of transfection, the cells were harvested for the further assays.
2.4 Total RNA extraction and quantitative reverse transcription-polymerase chain reaction
Total RNAs were obtained from cells through extraction applying the TRIzol reagent (Invitrogen). Then, NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific) was applied to examine the RNA concentration as well as purity. The complementary RNA was produced from total RNA employing the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA) (for miRNA), or the First Strand cDNA Synthesis Kit (Takara, Tokyo, Japan) (for mRNA). Quantitative real-time PCR was implemented on the 7500 Real-Time PCR System (Applied Biosystems) utilizing SYBR Green PCR Master Mix reagents (Takara, Otsu, Japan). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as normalized controls for mRNA and miRNA, respectively. was used for the determination of relative quantification. The primers were: miR-4319 forward, 5'-GCACAGCTCCCTGAGCAA-3', and reverse, 5'-CAGTGCGTGTCGTGGACT-3'; SMURF1 forward, 5'- CTGGATGCTTTTGGTCTGGT-3', and reverse, 5'- CCTGATAGACGCGAACACAG-3'; FUS forward, 5'- CTCCGGGAAACTGTGGCGTG-3', and reverse, 5'- ACAAAGTGGTCGTTGAGGGCA-3'; U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse, 5′-TTCACGAATTTGCGTGTCAT-3′; and GAPDH forward, 5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse, 5′-AGTCCTTCCACGATACCAAAGT-3′.
2.5 Cell Counting Kit-8 assay
The proliferative capacity of cells was determined with the application of Cell Counting Kit-8 (CCK-8; Dojindo, Japan). BHP5-16 and TPC cells were plated in the 96-well dish at a density of 5 × 104 cells in each well. After culturing cells for 0, 24, 48, 72, and 96 hours, each well was added with 10 μL of CCK-8. The measurement of absorbance (450 nm) was accomplished under a microplate reader (BioTek Instruments, Inc, Winooski, VT).
2.6 Transwell migration assay
The migration of BHP5-16 and TPC cells was detected utilizing 8 µm pore Transwell chambers (BD Biosciences, San Jose, CA). After transfection, 2 × 105 BHP5-16 and TPC cells were grown in non-serum medium (200 µL) and were plated in the upper insert. The lower insert was supplemented with 10% FBS. Subsequent to the incubation for about 2 days, the cells remaining in the upper insert were scrapped off with the use of a cotton swab. Then, the cells that had migrated to the bottom of membrane surface underwent fixation in 4% paraformaldehyde and staining using 1% crystal violet (Sigma-Aldrich Co, St Louis, MO). Migrated cells were calculated in five randomly chosen fields under an inverted microscope (Leica, Malvern, PA).
2.7 Luciferase reporter assay
The 3′UTR sections of SMURF1 containing the binding site for miR-4319 or the mutant site were amplified through polymerase chain reaction and were integrated into the psiCHECK2 vector (Promega, Madison, WI), named WT-SMURF1 and Mut-SMURF1. The WT-SMURF1 or Mut-SMURF1 reporters were transfected with miR-4319 mimic or NC mimic into 293T cells (American Type Culture Collection) with the aid of Lipofectamine 3000 (Thermo Fisher Scientific, Inc). The examination of luciferase activities was accomplished by the Dual Luciferase Reporter Assay System (Promega).
2.8 RNA immunoprecipitation
RNA immunoprecipitation (RIP) assay was conducted to evaluate the interaction of SMURF1 mRNA with miR-4319 and FUS with the utilization of the EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA). The antibodies including anti-Ago2 (ab32381; Abcam, Cambridge, UK), anti-FUS (ab23439; Abcam), and anti-immunoglobulin G (ab205719; Abcam) were from Abcam. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed to evaluate the levels of precipitated miR-4319 and SMURF1 mRNA.
2.9 Western blot analysis
The extracts of proteins were obtained utilizing the radioimmunoprecipitation assay lysis buffer (Beyotime, China). The examination of protein density was accomplished by the bicinchoninic acid protein assay (Pierce, Appleton). Twenty to thirty micrograms of proteins were loaded with the utilization of 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The loaded proteins were transferred to polyvinylidene fluoride membranes (Millipore). Subsequent to 1-hour sealing in 5% nonfat milk, the membranes were detected using primary antibodies for about 12 hours at 4℃, followed by culturing with a horseradish peroxidase-conjugated secondary antibody for another hour. The detection of signals was performed on the EasyBlot ECL detection system (Sangon, China). All primary antibodies, E-cadherin (ab40772; Abcam), N-cadherin (ab98953; Abcam), SMURF1 (ab57573; Abcam), and GAPDH (ab8245; Abcam), were from Abcam.
2.10 Statistical analysis
The data processing was conducted on the SPSS17.0 statistical software (SPSS Inc., Chicago, IL). Data presentation was performed by the mean ± standard deviation of results. The expression correlation was examined by Spearman's correlation analysis. All experiments were performed independently in triplicate. The comparison between applicable means was conducted using the Student t test, whereas the comparison of multiple groups was determined by one-way analysis of variance. The statistical significance was determined by P < .05.
2.11 Ethics Statement
All the patients had signed the written informed consents before enrollment, and the study had received the approval of the Ethics Committee of Xintai City People's Hospital of Shandong Province.
3 RESULTS
3.1 miR-4319 was downregulated in thyroid cancer, and overexpression of miR-4319 inhibited proliferation, migration, and EMT in thyroid cancer
First, we detected the participation of miR-4319 in thyroid cancer. We observed that miR-4319 expression level was lower in thyroid cancer tissues (n = 56) than in the matched adjacent noncancerous tissues (n = 56) (Figure 1A). By analyzing the correlation of miR-4319 level with clinical features, we confirmed that miR-4319 expression was associated with tumor size (P = .005), lymph node metastasis (P = .003), and clinical stage (P = .023) (Table 1). In addition, miR-4319 was downregulated in thyroid cancer cells compared with the normal cell line (Figure 1B).
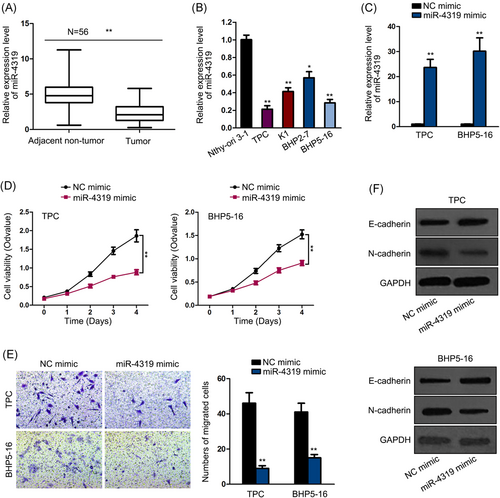
miR-4319 was downregulated in thyroid cancer and overexpression of miR-4319 inhibited proliferation, migration and EMT in thyroid cancer. A, Downregulation of miR-4319 in thyroid cancer tissues was confirmed by qPCR. B, Downregulation of miR-4319 in thyroid cancer cells was confirmed by qPCR. C, Transfection of miR-4319 mimic or NC mimic into TPC or BHP5-16 cells was confirmed by qPCR. D, CCK-8 assay was performed to examine the cell viability in response to miR-4319 overexpression. E, Transwell assay was implemented to evaluate cell migration. F, Western blot analysis was conducted to assess the levels of E-cadherin and N-cadherin. *P < .05, **P < .01. CCK-8, Cell Counting Kit-8; EMT, epithelial-to-mesenchymal transition; NC, negative control; qPCR, quantitative polymerase chain reaction
| Variable | miR-4319 expression | P value | |
|---|---|---|---|
| low | high | ||
| Age | |||
| <50 | 12 | 14 | .789 |
| ≥50 | 16 | 14 | |
| Sex | |||
| Female | 14 | 17 | .591 |
| Male | 14 | 11 | |
| Tumor Size | |||
| <3 cm | 5 | 16 | .005* |
| ≥3 cm | 23 | 12 | |
| Lymph node metastasis | |||
| No | 7 | 19 | .003* |
| Yes | 21 | 9 | |
| Distance metastasis | |||
| No | 18 | 20 | .775 |
| Yes | 10 | 8 | |
| Clinical stage | |||
| I/II | 5 | 14 | .023* |
| III/IV | 23 | 14 | |
| Pathological | |||
| Classical | 14 | 12 | .789 |
| Follicular | 14 | 16 | |
- Note: Low/high by the sample median. Pearson's χ2 test.
- * P < .05 was viewed as statistically significant.
Next, we further explored the function of miR-4319 in thyroid cancer through gain-of-function assays. We overexpressed the expression of miR-4319 in two thyroid cancer cell lines, TPC and BHP5-16, which were detected to present the lowest miR-4319 levels. The marked overexpression of miR-4319 in two cell lines after miR-4319 mimic transfection was verified by quantitative polymerase chain reaction (qPCR) results (Figure 1C). Subsequently, we observed the attenuated cell proliferation upon miR-4319 overexpression in thyroid cancer cells (Figure 1D). Transwell assay demonstrated the decreased number of migrated cells in response to miR-4319 overexpression in thyroid cancer cells (Figure 1E). In addition, EMT markers were tested. Western blot analysis showed that ectopic miR-4319 expression induced the E-cadherin level and reduced N-cadherin level (Figure 1F). In summary, it was indicated that miR-4319 was downregulated in thyroid cancer, and miR-4319 overexpression repressed cell proliferation, migration, and EMT progression.
3.2 miR-4319 targeted SMURF1 in thyroid cancer cells
Thereafter, we interrogated the mechanism whereby miR-4319 regulated thyroid cancer. miRNAs are commonly known as suppressors of gene expression in cancers by targeting the 3′UTR region of certain mRNAs. By browsing the miRDB (http://mirdb.org/cgi-bin/target_detail.cgi?targetID = 1480874), we identified SMURF1 as a potent target gene for miR-4319. SMURF1 has been illustrated to promote proliferation, migration, and EMT in diverse cancers21, 22 and has also been newly discovered to promote thyroid cancer progression.23 Therefore, we focused on investigating SMURF1. The binding sites on the SMURF1 3′UTR region for miR-4319 and mutant sites are shown in Figure 2A. The results of qPCR and Western blot analyses showed the upregulation of SMURF1 expression in thyroid cancer tissues at mRNA and protein levels (Figure 2B). Also, SMURF1 mRNA and protein expressions were upregulated in thyroid cancer cell lines compared with a normal cell line (Figure 2C). Spearman's correlation curve presented the negative correlation between miR-4319 and SMURF1 expressions in thyroid cancer tissues (Figure 2D). The RIP assay followed by qPCR analysis validated that miR-4319 and SMURF1 mRNA could be co-immunoprecipitated by anti-Ago2 (Figure 2E), confirming the interaction between miR-4319 and SMURF1 mRNA. Moreover, the overexpression of miR-4319 alleviated the luciferase activity on WT-SMURF1 instead of Mut-SMURF1 (Figure 2F). Furthermore, ectopic expression of miR-4319 decreased mRNA and protein levels of SMURF1 in thyroid cancer cells (Figure 2G,H). The results collectively implied that miR-4319 targeted SMURF1 in thyroid cancer cells.
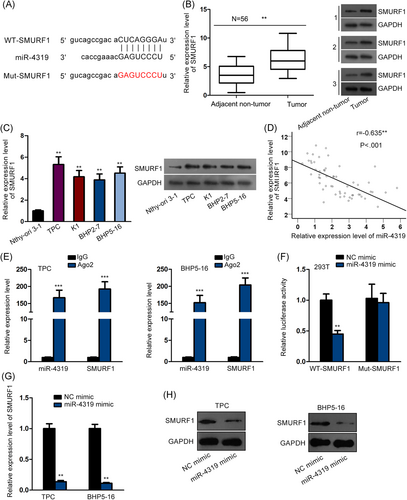
miR-4319 targeted SMURF1 in thyroid cancer cells. A, The binding sequences between SMURF1 and miR-4319 and the mutant sequences on SMURF1. B, The upregulation of SMURF1 in thyroid cancer tissues was verified by qPCR and Western blot analyses. C, The upregulation of SMURF1 in thyroid cancer cells was verified by qPCR and Western blot analyses. D, Spearman's correlation analysis confirmed the negative correlation between miR-4319 and SMURF1 expression in thyroid cancer tissues. E, RIP assay was use to evaluate the interaction between miR-4319 and SMURF1. F, Luciferase reporter assay was used to assess the targeting of miR-4319 on SMURF1 3′UTR. G,H, The effect of miR-4319 on the mRNA and protein levels of SMURF1 was assessed by qPCR and Western blot. **P < .01, ***P < .001. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IgG, immunoglobulin G; NC, negative control; qPCR, quantitative polymerase chain reaction; UTR, untranslated region
3.3 miR-4319 inhibited the stabilization of SMURF1 mRNA by competing with FUS to target SMURF1
Furthermore, through browsing Starbase (http://starbase.sysu.edu.cn/starbase2/index.php), we found that FUS was a potential interacting protein with SMURF1. FUS is an RNA-binding protein with multiple functions, including transcription activation in nucleus and mRNA stabilization in cytoplasm.15, 16 In addition, FUS has been proved to participate in thyroid cancer.17 Therefore, we further probed the influence of FUS on SMURF1. RIP assay results showed the abundance of SMURF1 mRNA in the precipitates of anti-FUS (Figure 3A), confirming the interaction between FUS and SMURF1 mRNA. Thereafter, we detected the effect of FUS on SMURF1. The pronounced knockdown of FUS by shRNAs was confirmed by qPCR analysis (Figure 3B). Actinomycin D was then added to inhibit new mRNA production in thyroid cancer cells. The qPCR analysis of the SMURF1 mRNA level was conducted over time, and the results showed that silencing FUS shortened the half-life of SMURF1 mRNA (Figure 3C), indicating FUS as an mRNA stabilizer for SMURF1. Recent studies pointed out that miRNAs could either coordinate or interfere with RBPs to regulate their shared target mRNAs,13, 14 so we tried to test whether miR-4319 could interfere with the regulation of FUS on SMURF1. The RIP assay showed that overexpressing miR-4319 reduced the expression of SMURF1 mRNA in the precipitates of anti-FUS (Figure 3D). Furthermore, we validated that overexpressing miR-4319 had no effect on the expression of FUS (Figure 3E). The results of qPCR and Western blot analyses showed that overexpressing FUS rescued the expressions of SMURF1 mRNA and protein, which were reduced by miR-4319 (Figure 3F,G). Thus, the results above suggested that miR-4319 inhibited the stabilization of FUS on SMURF1 mRNA by competing with FUS to target SMURF1.
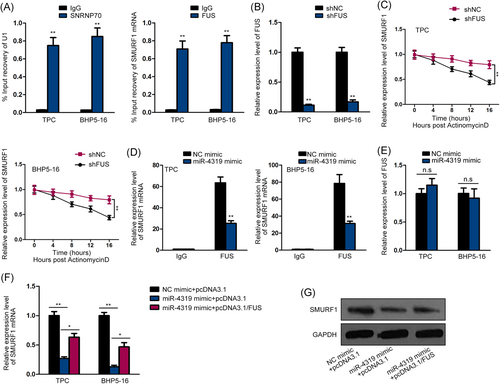
miR-4319 inhibited the stabilization of SMURF1 mRNA by competing with FUS to target SMURF1. A, RIP assay was used to evaluate the interplay between FUS and SMURF1. SNRNP70 and U1 were used as positive control. B, The knockdown of FUS by shRNAs was confirmed by qPCR. C, Time course experiments were used to examine the effect of FUS on the mRNA stability of SMURF1. D, RIP assay was used to examine the effect of miR-4319 on the interaction between FUS and SMURF1. E, qPCR analysis confirmed that miR-4319 had no effect on the expression of FUS. F,G, qPCR and Western blot analyses showed that overexpression of FUS could abrogate the inhibitive effect of miR-4319 upregulation on SMURF1 mRNA and protein levels. *P < .05, **P < .01, (n.s, no significance). FUS, fused in sarcoma; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IgG, immunoglobulin G; mRNA, messenger RNA; NC, negative control; qPCR, quantitative polymerase chain reaction; RIP, RNA immunoprecipitation; shRNAs, short hairpin RNAs;
3.4 miR-4319 inhibited cell proliferation, migration, and EMT through SMURF1 in thyroid cancer
Finally, we unfolded the rescue experiments to investigate whether miR-4319 inhibited thyroid cancer progression through SMURF1. The qPCR analysis confirmed that overexpressing SMURF1 restored SMURF1 expression decreased by miR-4319 overexpression (Figure 4A). The CCK-8 assay demonstrated that overexpressing SMURF1 recovered the cell proliferation attenuated by miR-4319 overexpression in thyroid cancer cells (Figure 4B). Also, cotransfection of pcDNA3.1/SMURF1 impaired the inhibitive effect of miR-4319 overexpression on cell migration in thyroid cancer cells (Figure 4C). The results of Western blot analysis depicted that the increase of E-cadherin and decrease of N-cadherin expression caused by miR-4319 overexpression were reversed by overexpressing SMURF1 (Figure 4D). These results implied that miR-4319 inhibited cell proliferation, migration, and EMT through SMURF1 in thyroid cancer.
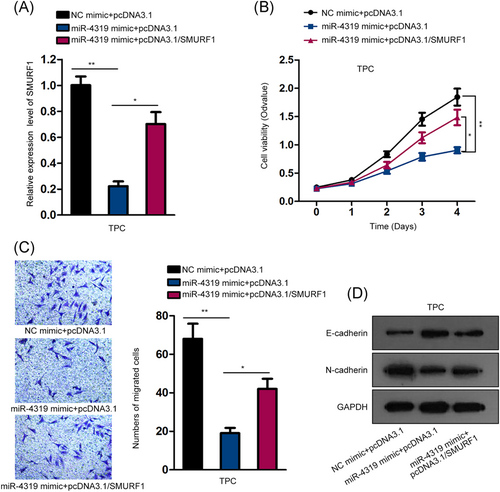
miR-4319 inhibited cell proliferation, migration, and EMT through SMURF1 in thyroid cancer. A, qPCR analysis confirmed that overexpressing SMURF1 impaired the inhibitory effect of miR-4319 overexpression on SMURF1 mRNA level. B, CCK-8 assay was used to evaluate cell viability in each group. C, Transwell migration assay was used to assess cell migration in each group. D Western blot analysis was performed to determine the levels of E-cadherin and N-cadherin in each group. *P < .05, **P < .01. CCK-8, Cell Counting Kit-8; EMT, epithelial-to-mesenchymal transition; mRNA, messenger RNA; NC, negative control; qPCR, quantitative polymerase chain reaction
4 DISCUSSION
Over the past few decades, the morbidity and deaths due to thyroid cancer has been rising.2, 3 The carcinogenesis and tumor progression of thyroid cancer result from varieties of factors, including variation of gene expression and changing activity in signaling pathways.4, 5 Dysregulation of miRNAs has been documented to be implicated in a number of cancers,8, 9 including thyroid cancer.10 Therefore, the exploration on miRNAs in thyroid cancer is helpful for the identification of potential biological markers for thyroid cancer.
Previous studies elucidated that miR-4319 inhibited tumor progression in breast cancer and acute myeloid leukemia.11, 12 Accordingly, our study first showed that miR-4319 exhibited low expression in thyroid cancer tissues and cell lines. In addition, we revealed the correlation of miR-4319 expression with tumor size, lymph node metastasis, and clinical stage in thyroid cancer patients, indicating that miR-4319 might participate in thyroid cancer. Gain-of-function assays suggested that miR-4319 restrained proliferation, migration, and EMT progression in thyroid cancer cells.
The major mechanism whereby miRNAs administrate gene expression is directly targeting the 3′UTR of mRNAs.7 In the current study, we searched miRDB and found that SMURF1 was a potential target for miR-4319. SMURF1 is known to be a C2-WW-HECT ubiquitin ligase, which can modulate multiple biological activities, including viral autophagy embryogenesis, and bone homeostasis.18-20 Previously, studies have illustrated that SMURF1 can accelerate proliferation, migration, and EMT in diverse cancers.21, 22 More importantly, it has been proved that SMURF1 plays a carcinogenic role in thyroid cancer.23 In concordance, we verified the high expression of SMURF1 in thyroid cancer tissues and cells, and showed the negative correlation between miR-4319 and SMURF1 in thyroid cancer tissues. Also, we were the first to prove that SMURF1 was targeted by miR-4319 in thyroid cancer.
Recent studies illustrated that miRNAs could coordinate or interfere with RBPs to regulate their shared target mRNAs.13, 14 Based on the prediction results of the Starbase, we found that FUS potentially interacted with SMURF1. FUS is an RNA-binding protein that can regulate transcription activation in the nucleus and mRNA stabilization in cytoplasm.15, 16 Our study first revealed that FUS stabilized SMURF1 mRNA in thyroid cancer cells, and that miR-4319 inhibited the stabilization of SMURF1 mRNA by FUS. Finally, rescue assays indicated that miR-4319 repressed proliferation, migration, and EMT through SMURF1 in thyroid cancer.
In sum, the current study illustrated that miR-4319 inhibited the development of thyroid cancer by modulating FUS-stabilized SMURF1, presenting miR-4319 as a new biological target for thyroid cancer.
ACKNOWLEDGMENTS
The author would like to thank all the people involved in this study.
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interests.
AUTHOR CONTRIBUTION
SB contributed to article writing and experiment preparation as well as data collection.